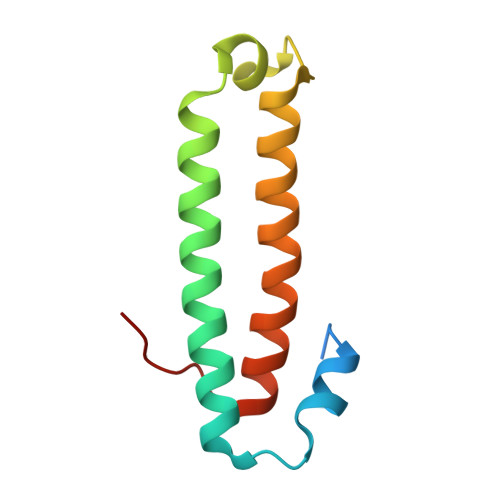
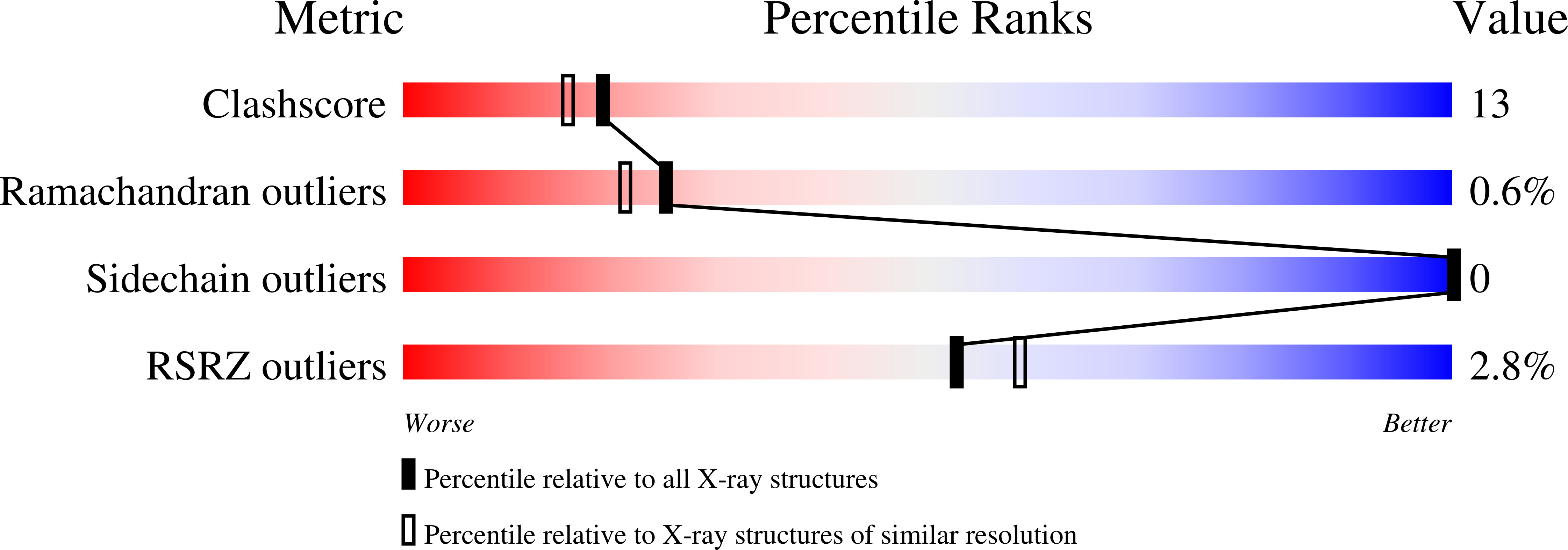Crystal Structure and Mechanism of TraM2, a Second Quorum-Sensing Antiactivator of Agrobacterium tumefaciens Strain A6.
Chen, G., Wang, C., Fuqua, C., Zhang, L.H., Chen, L.(2006) J Bacteriol 188: 8244-8251
- PubMed: 16997969
- DOI: https://doi.org/10.1128/JB.00954-06
- Primary Citation of Related Structures:
2HJD - PubMed Abstract:
Quorum sensing is a community behavior that bacteria utilize to coordinate a variety of population density-dependent biological functions. In Agrobacterium tumefaciens, quorum sensing regulates the replication and conjugative transfer of the tumor-inducing (Ti) plasmid from pathogenic strains to nonpathogenic derivatives. Most of the quorum-sensing regulatory proteins are encoded within the Ti plasmid. Among these, TraR is a LuxR-type transcription factor playing a key role as the quorum-sensing signal receptor, and TraM is an antiactivator that antagonizes TraR through the formation of a stable oligomeric complex. Recently, a second TraM homologue called TraM2, not encoded on the Ti plasmid of A. tumefaciens A6, was identified, in addition to a copy on the Ti plasmid. In this report, we have characterized TraM2 and its interaction with TraR and solved its crystal structure to 2.1 A. Like TraM, TraM2 folds into a helical bundle and exists as homodimer. TraM2 forms a stable complex (K(d) = 8.6 nM) with TraR in a 1:1 binding ratio, a weaker affinity than that of TraM for TraR. Structural analysis and biochemical studies suggest that protein stability may account for the difference between TraM2 and TraM in their binding affinities to TraR and provide a structural basis for L54 in promoting structural stability of TraM.
Organizational Affiliation:
Department of Biology, 915 E. 3rd St., Indiana University, Bloomington, Indiana 47405, USA.