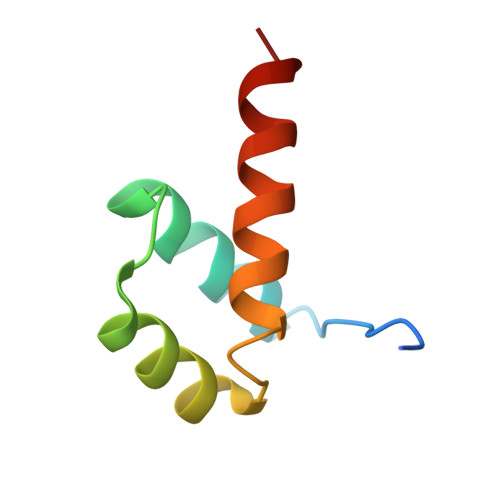
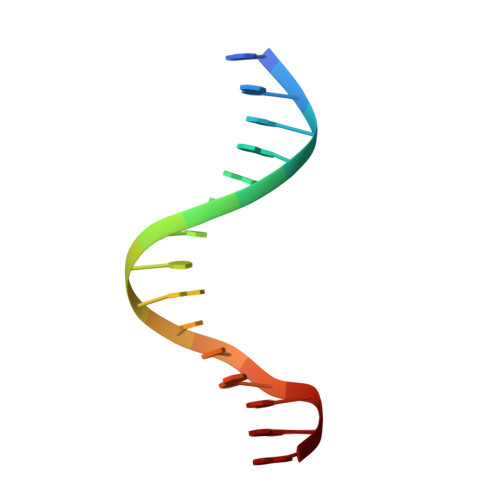
Structural basis for induced fit mechanisms in DNA recognition by the pdx1 homeodomain
Longo, A., Guanga, G.P., Rose, R.B.(2007) Biochemistry 46: 2948-2957
- PubMed: 17315980
- DOI: https://doi.org/10.1021/bi060969l
- Primary Citation of Related Structures:
2H1K - PubMed Abstract:
Pancreatic and duodenal homeobox 1 (Pdx1) is a homeodomain transcription factor belonging to the ParaHox family. Pdx1 plays an essential role in pancreatic endocrine and exocrine cell development and maintenance of adult islet beta-cell function. Mutations in the human pdx1 gene are linked to an early onset form of non-insulin-dependent diabetes mellitus, MODY-4. We demonstrate that the homeodomain reproduces the binding specificity of the full-length protein. We report the 2.4 A resolution crystal structure of the homeodomain bound to a target DNA. The two Pdx1/DNA complexes in the asymmetric unit display conformational differences: in the DNA curvature, the orientation of the homeodomain in the major groove, and the order of the N-terminal arm. Comparing the two complexes indicates invariant protein-DNA contacts, and variant contacts that are unique to each binding orientation. An induced fit model is proposed that depends on the DNA conformation and provides a mechanism for nonlocal contributions to binding specificity.
Organizational Affiliation:
Department of Molecular and Structural Biochemistry, 128 Polk Hall, North Carolina State University, Raleigh, North Carolina 27695, USA.