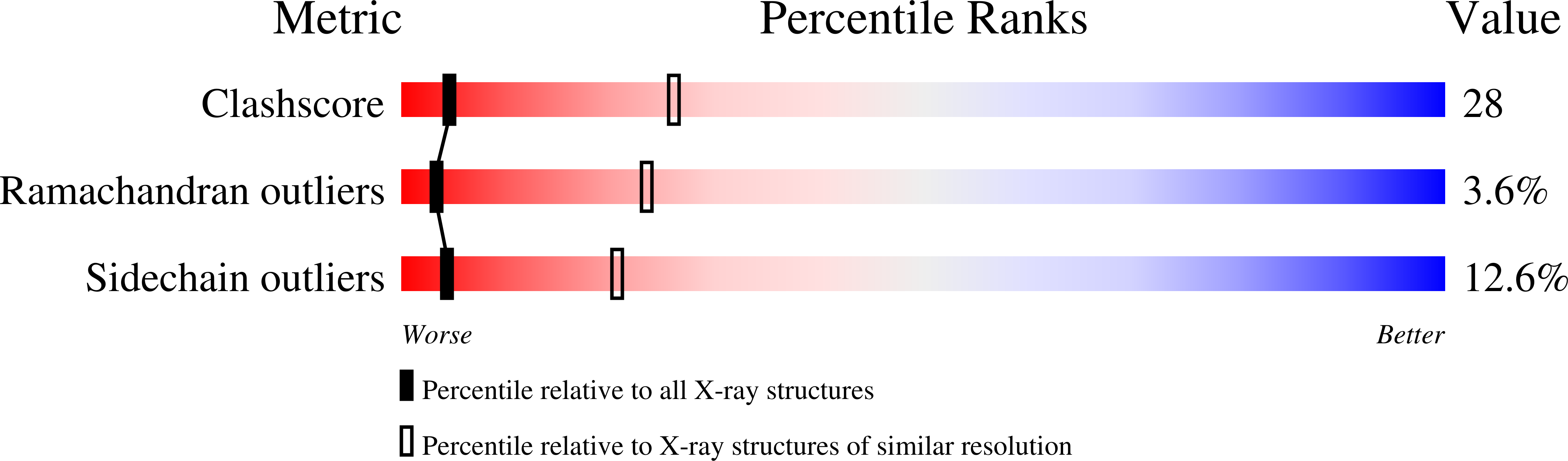Structurally mapping the diverse phenotype of adeno-associated virus serotype 4.
Govindasamy, L., Padron, E., McKenna, R., Muzyczka, N., Kaludov, N., Chiorini, J.A., Agbandje-McKenna, M.(2006) J Virol 80: 11556-11570
- PubMed: 16971437
- DOI: https://doi.org/10.1128/JVI.01536-06
- Primary Citation of Related Structures:
2G8G - PubMed Abstract:
The adeno-associated viruses (AAVs) can package and deliver foreign DNA into cells for corrective gene delivery applications. The AAV serotypes have distinct cell binding, transduction, and antigenic characteristics that have been shown to be dictated by the capsid viral protein (VP) sequence. To understand the contribution of capsid structure to these properties, we have determined the crystal structure of AAV serotype 4 (AAV4), one of the most diverse serotypes with respect to capsid protein sequence and antigenic reactivity. Structural comparison of AAV4 to AAV2 shows conservation of the core beta strands (betaB to betaI) and helical (alphaA) secondary structure elements, which also exist in all other known parvovirus structures. However, surface loop variations (I to IX), some containing compensating structural insertions and deletions in adjacent regions, result in local topological differences on the capsid surface. These include AAV4 having a deeper twofold depression, wider and rounder protrusions surrounding the threefold axes, and a different topology at the top of the fivefold channel from that of AAV2. Also, the previously observed "valleys" between the threefold protrusions, containing AAV2's heparin binding residues, are narrower in AAV4. The observed differences in loop topologies at subunit interfaces are consistent with the inability of AAV2 and AAV4 VPs to combine for mosaic capsid formation in efforts to engineer novel tropisms. Significantly, all of the surface loop variations are associated with amino acids reported to affect receptor recognition, transduction, and anticapsid antibody reactivity for AAV2. This observation suggests that these capsid regions may also play similar roles in the other AAV serotypes.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Center for Structural Biology, The McKnight Brain Institute, College of Medicine, University of Florida, Gainesville, FL 32610, USA.