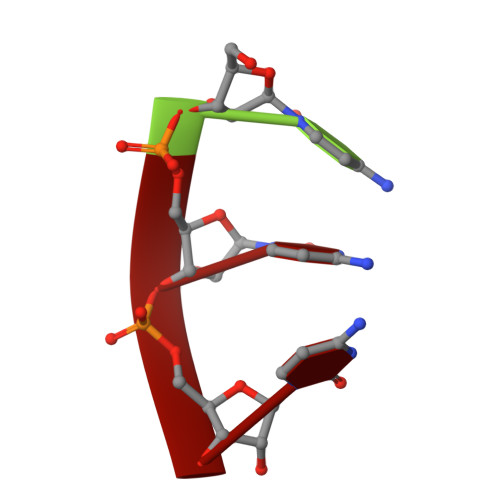
The RNA of turnip yellow mosaic virus exhibits icosahedral order.
Larson, S.B., Lucas, R.W., Greenwood, A., McPherson, A.(2005) Virology 334: 245-254
- PubMed: 15780874
- DOI: https://doi.org/10.1016/j.virol.2005.01.036
- Primary Citation of Related Structures:
2FZ1, 2FZ2 - PubMed Abstract:
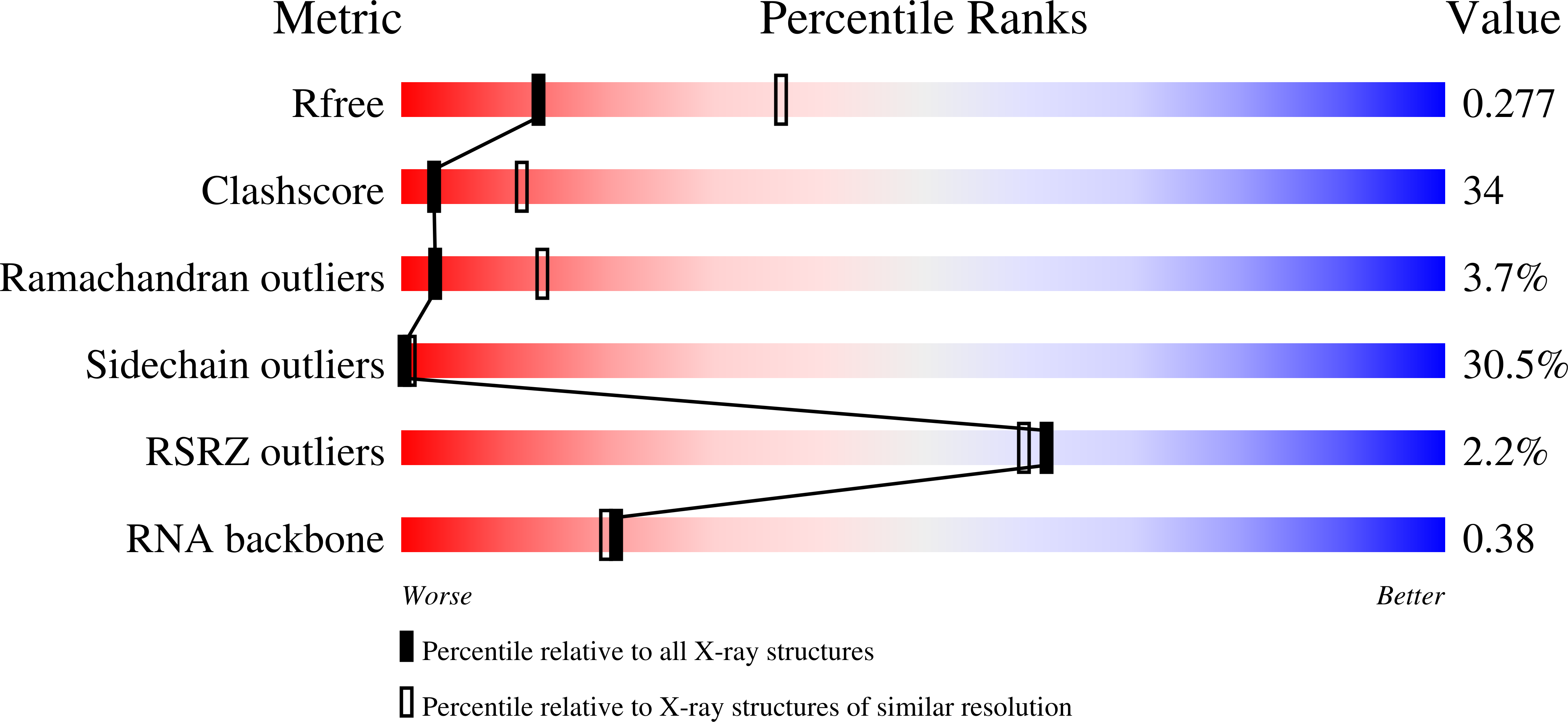
Difference electron density maps, based on structure factor amplitudes and experimental phases from crystals of wild-type turnip yellow mosaic virus and those of empty capsids prepared by freeze-thawing, show a large portion of the encapsidated RNA to have an icosahedral distribution. Four unique segments of base-paired, double-helical RNA, one to two turns in length, lie between 33-A and 101-A radius and are organized about either 2-fold or 5-fold icosahedral axes. In addition, single-stranded loops of RNA invade the pentameric and hexameric capsomeres where they contact the interior capsid surface. The remaining RNA, not seen in electron density maps, must serve as connecting links between these secondary structural elements and is likely icosahedrally disordered. The distribution of RNA observed crystallographically appears to be in agreement with models based on biochemical data and secondary structural analyses.
Organizational Affiliation:
Department of Molecular Biology and Biochemistry, University of California, Irvine, CA 92697-3900, USA.