Solution Structure and Interactions of the Escherichia Coli Cell Division Activator Protein Ceda.
Chen, H.A., Simpson, P., Huyton, T., Roper, D., Matthews, S.(2005) Biochemistry 44: 6738
- PubMed: 15865419
- DOI: https://doi.org/10.1021/bi0500269
- Primary Citation of Related Structures:
2BN8 - PubMed Abstract:
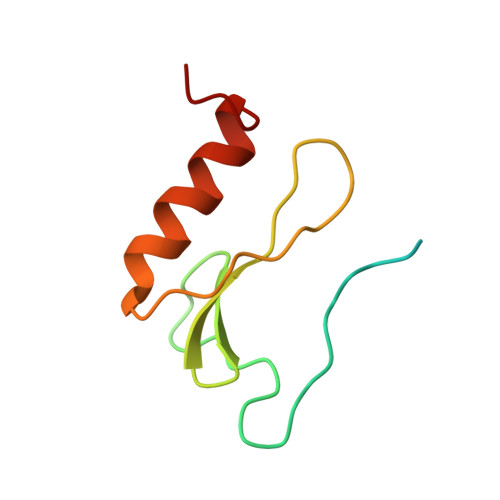
CedA is a protein that is postulated to be involved in the regulation of cell division in Escherichia coli and related organisms; however, little biological data about its possible mode of action are available. Here we present a three-dimensional structure of this protein as determined by NMR spectroscopy. The protein is made up of four antiparallel beta-strands, an alpha-helix, and a large unstructured stretch of residues at the N-terminus. It shows structural similarity to a family of DNA-binding proteins which interact with dsDNA via a three-stranded beta-sheet, suggesting that CedA may be a DNA-binding protein. The putative binding surface of CedA is predominantly positively charged with a number of basic residues surrounding a groove largely dominated by aromatic residues. NMR chemical shift perturbations and gel-shift experiments performed with CedA confirm that the protein binds dsDNA, and its interaction is mediated primarily via the beta-sheet.
Organizational Affiliation:
Department of Biological Sciences and Centre for Structural Biology, Imperial College of Science, Technology and Medicine, South Kensington, London, UK.