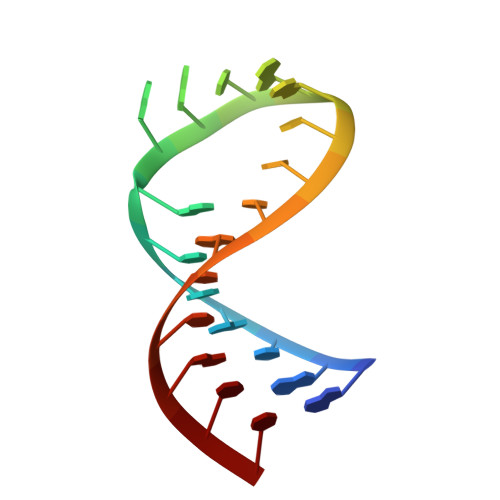
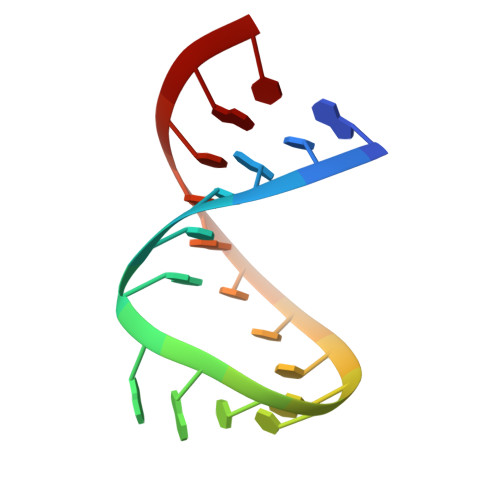
The solution structure of an RNA loop-loop complex: the ColE1 inverted loop sequence.
Lee, A.J., Crothers, D.M.(1998) Structure 6: 993-1005
- PubMed: 9739090
- DOI: https://doi.org/10.1016/s0969-2126(98)00101-4
- Primary Citation of Related Structures:
1BJ2, 2BJ2 - PubMed Abstract:
Replication of the ColE1 plasmid of Escherichia coli is regulated by the interaction of sense and antisense plasmid-encoded transcripts. The antisense RNA I negatively regulates the replication of the plasmid by duplex formation with complementary RNA II. The interaction is initiated by the formation of a double helix between seven-nucleotide loops from each RNA and is stabilized by binding of the RNA one modulator (ROM) protein. The ROM protein is thought to recognize a specific RNA structure, regardless of sequence. The solution structure of a loop-loop complex between model RNA hairpins that resemble RNA I and RNA II has been determined by nuclear magnetic resonance spectroscopy. The model hairpins have loop sequences inverted 5' to 3' relative to the wild-type sequence and were chosen because of their complex's slow dissociation in comparison to the wild type. The complex has continuous stacking from the 3'-side of one stem helix through the loop-loop helix to the other stem helix. One residue from each hairpin has a unique phosphodiester bond which bridges and narrows the major groove. These bridging phosphates are in close proximity to the phosphate groups of the adjacent bases, forming unique structural motifs called phosphate clusters. The purine residue at the 3'-end of the loop-loop helix of one RNA stacks on a purine residue on the 5'-side of the other RNA stem, and there are strong cross-strand stacking interactions between guanine bases in the stem helices adjacent to the loops. Unique distortions, such as the strong bend and the phosphate clusters flanking the major groove of the loop-loop helix, provide an attractive nonsequence-specific structural feature for recognition by the ROM protein. The structure provides a basis for rationalizing the sequence dependence of the stability of loop-loop interaction.
Organizational Affiliation:
Department of Chemistry, Yale University, New Haven, CT 06520-8107, USA.