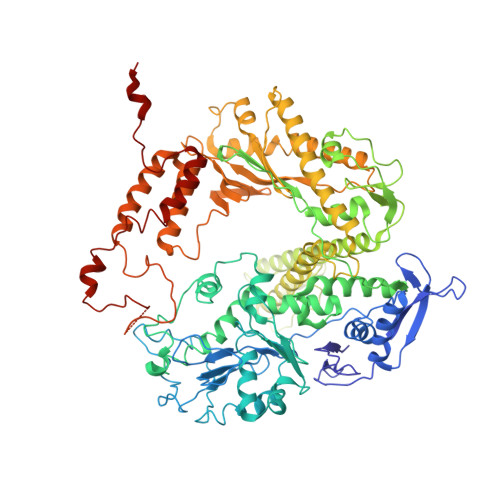
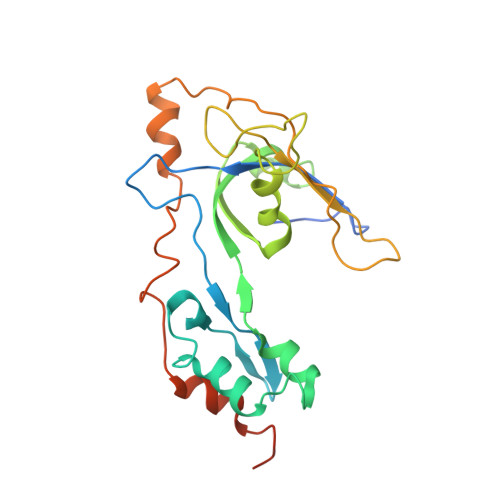
Structure and enzymatic properties of a chimeric bacteriophage RB69 DNA polymerase and single-stranded DNA binding protein with increased processivity.
Sun, S., Geng, L., Shamoo, Y.(2006) Proteins 65: 231-238
- PubMed: 16881051
- DOI: https://doi.org/10.1002/prot.21088
- Primary Citation of Related Structures:
2A1K, 2ATQ - PubMed Abstract:
In vivo, replicative DNA polymerases are made more processive by their interactions with accessory proteins at the replication fork. Single-stranded DNA binding protein (SSB) is an essential protein that binds tightly and cooperatively to single-stranded DNA during replication to remove adventitious secondary structures and protect the exposed DNA from endogenous nucleases. Using information from high resolution structures and biochemical data, we have engineered a functional chimeric enzyme of the bacteriophage RB69 DNA polymerase and SSB with substantially increased processivity. Fusion of RB69 DNA polymerase with its cognate SSB via a short six amino acid linker increases affinity for primer-template DNA by sixfold and subsequently increases processivity by sevenfold while maintaining fidelity. The crystal structure of this fusion protein was solved by a combination of multiwavelength anomalous diffraction and molecular replacement to 3.2 A resolution and shows that RB69 SSB is positioned proximal to the N-terminal domain of RB69 DNA polymerase near the template strand channel. The structural and biochemical data suggest that SSB interactions with DNA polymerase are transient and flexible, consistent with models of a dynamic replisome during elongation.
Organizational Affiliation:
Department of Biochemistry and Cell Biology, Rice University, Houston, Texas 77005, USA.