Structure of replicative DNA polymerase provides insigts into the mechanisms for processivity, frameshifting and editing
Brieba, L., Ellenberger, T.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| T7 DNA polymerase | E [auth A], G [auth F] | 704 | Escherichia phage T7 | Mutation(s): 2 Gene Names: 5 EC: 2.7.7.7 | 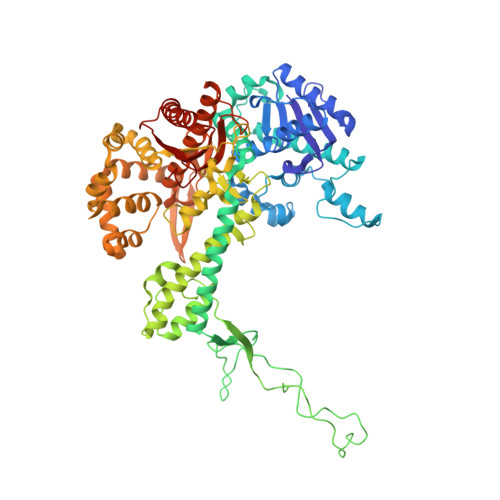 |
UniProt | |||||
Find proteins for P00581 (Escherichia phage T7) Explore P00581 Go to UniProtKB: P00581 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P00581 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| thioredoxin 1 | F [auth B], H [auth I] | 108 | Escherichia coli | Mutation(s): 0 Gene Names: trxA, fipA, tsnC | 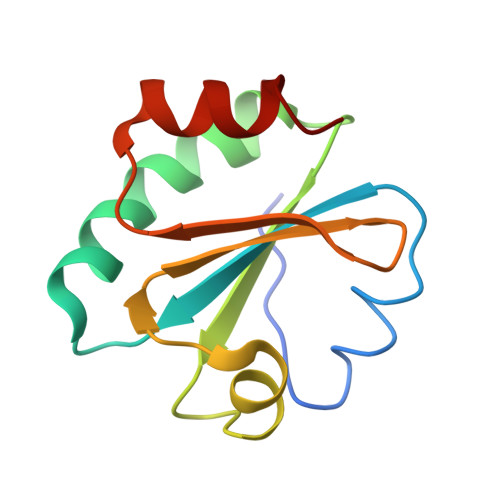 |
UniProt | |||||
Find proteins for P0AA25 (Escherichia coli (strain K12)) Explore P0AA25 Go to UniProtKB: P0AA25 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0AA25 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA Primer | A [auth P], C [auth X] | 22 | N/A | 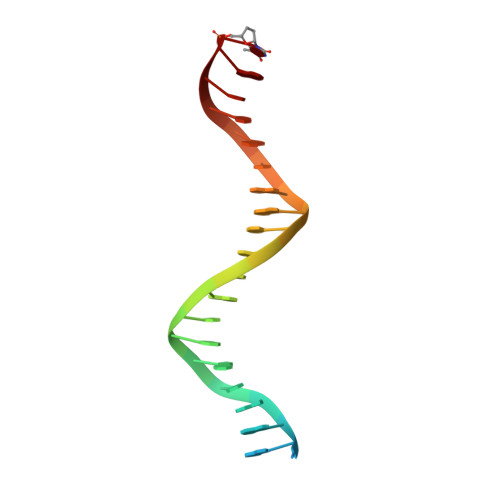 | |
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA Template | B [auth T], D [auth Z] | 26 | N/A | 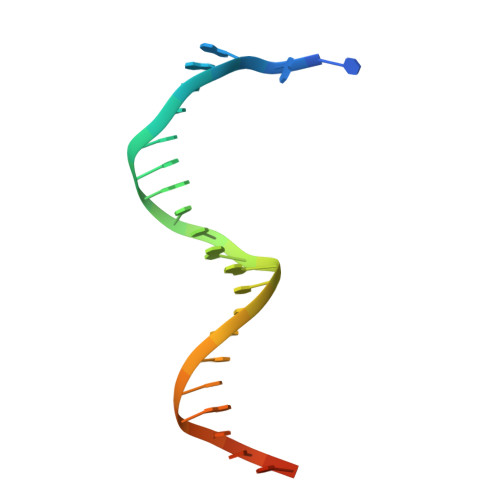 | |
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 168.312 | α = 90 |
| b = 169.235 | β = 90 |
| c = 179.788 | γ = 90 |
| Software Name | Purpose |
|---|---|
| HKL-2000 | data collection |
| TRUNCATE | data reduction |
| CNS | refinement |
| HKL-2000 | data reduction |
| CCP4 | data scaling |
| CNS | phasing |