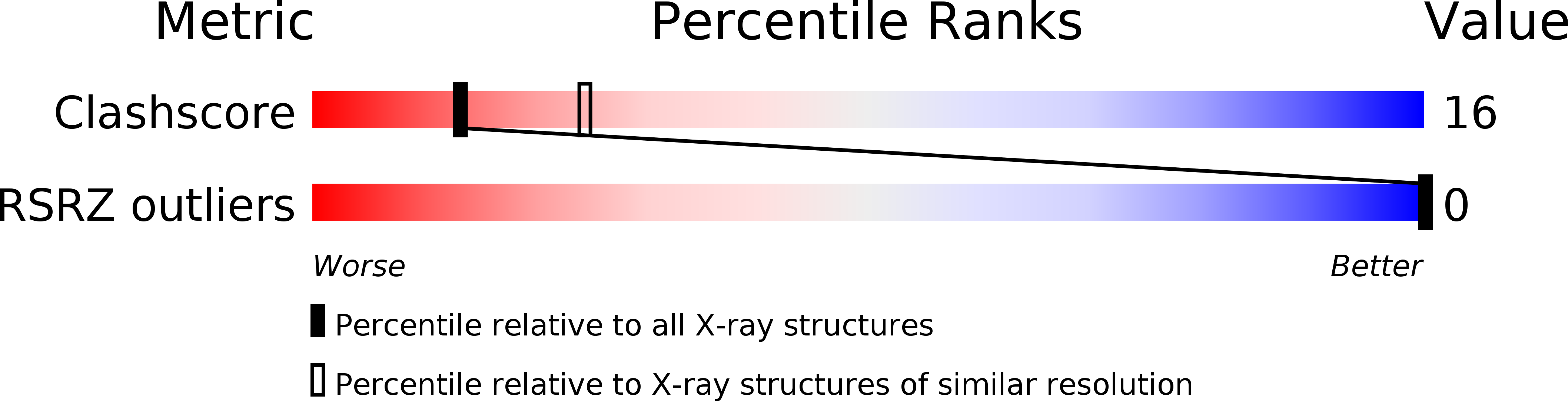X-ray and solution studies of DNA oligomers and implications for the structural basis of A-tract-dependent curvature.
Shatzky-Schwartz, M., Arbuckle, N.D., Eisenstein, M., Rabinovich, D., Bareket-Samish, A., Haran, T.E., Luisi, B.F., Shakked, Z.(1997) J Mol Biol 267: 595-623
- PubMed: 9126841
- DOI: https://doi.org/10.1006/jmbi.1996.0878
- Primary Citation of Related Structures:
285D, 286D, 287D, 297D - PubMed Abstract:
DNA containing short periodic stretches of adenine residues (known as A-tracts), which are aligned with the helical repeat, exhibit a pronounced macroscopic curvature. This property is thought to arise from the cumulative effects of a distinctive structure of the A-tract. It has also been observed by gel electrophoresis that macroscopic curvature is largely retained when inosine bases are introduced singly into A-tracts but decreases abruptly for pure I-tracts. The structural basis of this effect is unknown. Here we describe X-ray and gel electrophoretic analyses of several oligomers incorporating adenine or inosine bases or both. We find that macroscopic curvature is correlated with a distinctive base-stacking geometry characterized by propeller twisting of the base-pairs. Regions of alternating adenine and inosine bases display large propeller twisting comparable to that of pure A-tracts, whereas the values observed for pure I-tracts are significantly smaller. We also observe in the crystal structures that propeller twist leads to close cross-strand contacts between amino groups from adenine and cytosine bases, indicating an attractive NH-N interaction, which is analogous to the NH-O interaction proposed for A-tracts. This interaction also occurs between adenine bases across an A-T step and may explain in part the different behavior of A-T versus T-A steps in the context of A-tract-induced curvature. We also note that hydration patterns may contribute to propeller-twisted conformation. Based on the present data and other structural and biophysical studies, we propose that DNA macroscopic curvature is related to the structural invariance of A-tract and A-tract-like regions conferred by high propeller twist, cross-strand interactions and characteristic hydration. The implications of these findings to the mechanism of DNA bending are discussed.
Organizational Affiliation:
Department of Structural Biology, Weizmann Institute of Science, Rehovot, Israel.