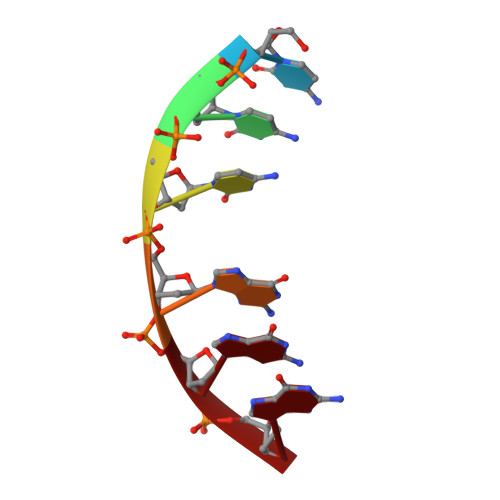
A continuous transition from A-DNA to B-DNA in the 1:1 complex between nogalamycin and the hexamer dCCCGGG.
Cruse, W.B., Saludjian, P., Leroux, Y., Leger, G., Manouni, D.E., Prange, T.(1996) J Biol Chem 271: 15558-15567
- PubMed: 8662899
- DOI: https://doi.org/10.1074/jbc.271.26.15558
- Primary Citation of Related Structures:
282D - PubMed Abstract:
The antibiotic nogalamycin, a drug with high specificity for TG and CG steps in double-stranded DNA, has been crystallized as a 1:1 complex with the hexamer d(CCCGGG). The antibiotic is inserted at the central CG step of the duplex, with the two sugars oriented in the same direction and with strong interactions with the DNA within the grooves. The amino-glucose residue makes an integral part of a well defined major groove hydration network with van der Waals contacts and several strong hydrogen bonds to the duplex. The nogalose residue resides in the minor groove, making primarily van der Waals contacts. The single site allows an accurate molecular description of the intercalation, without perturbations from end effects observed previously. The local unwinding induced by nogalamycin is completely relaxed 2 base pairs away from the intercalation site. The two strands of the DNA show a continuous deformation from the A to the B form: 1) the cytosines toward the 5' end of the nogalomycin site in each strand have c3'-endo conformations while 5 guanosines toward the 3' ends have c2'-endo conformations; 2) within each strand, the phosphate-phosphate distances increase in a continuous manner from 5.7 A (A-form) to 7.1 A (B-form).
Organizational Affiliation:
"Chimie Structurale et Spectroscopie Biomoléculaire" (URA 1430 CNRS) UFR-Biomédicale, 74, rue M. Cachin, 93012 Bobigny Cedex, France.