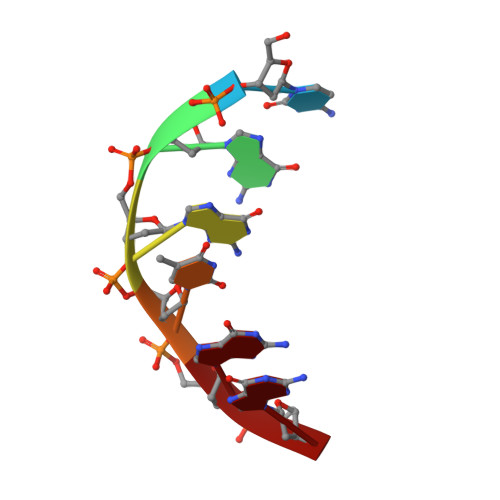
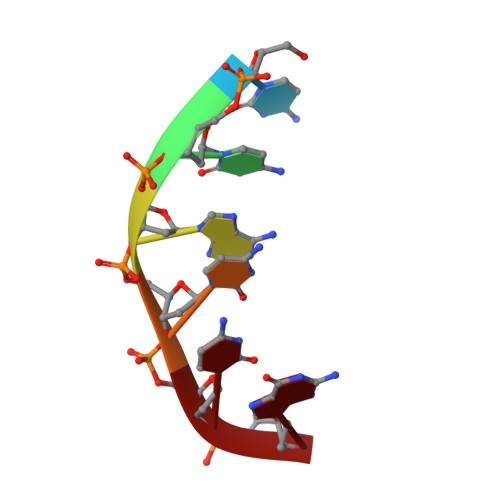
Base-pair opening and spermine binding--B-DNA features displayed in the crystal structure of a gal operon fragment: implications for protein-DNA recognition.
Tari, L.W., Secco, A.S.(1995) Nucleic Acids Res 23: 2065-2073
- PubMed: 7596838
- DOI: https://doi.org/10.1093/nar/23.11.2065
- Primary Citation of Related Structures:
206D - PubMed Abstract:
A sequence that is represented frequently in functionally important sites involving protein-DNA interactions is GTG/CAC, suggesting that the trimer may play a role in regulatory processes. The 2.5 A resolution structure of d(CGGTGG)/d(CCACCG), a part of the interior operator (OI, nucleotides +44 to +49) of the gal operon, co-crystallized with spermine, is described herein. The crystal packing arrangement in this structure is unprecedented in a crystal of B-DNA, revealing a close packing of columns of stacked DNA resembling a 5-stranded twisted wire cable. The final structure contains one hexamer duplex, 17 water molecules and 1.5 spermine molecules per crystallographic asymmetric unit. The hexamer exhibits base-pair opening and shearing at T.A resulting in a novel non-Watson-Crick hydrogen-bonding scheme between adenine and thymine in the GTG region. The ability of this sequence to adopt unusual conformations in its GTG region may be a critical factor conferring sequence selectivity on the binding of Gal repressor. In addition, this is the first conclusive example of a crystal structure of spermine with native B-DNA, providing insight into the mechanics of polyamine-DNA binding, as well as possible explanations for the biological action of spermine.
Organizational Affiliation:
Department of Chemistry, University of Manitoba, Winnipeg, Canada.