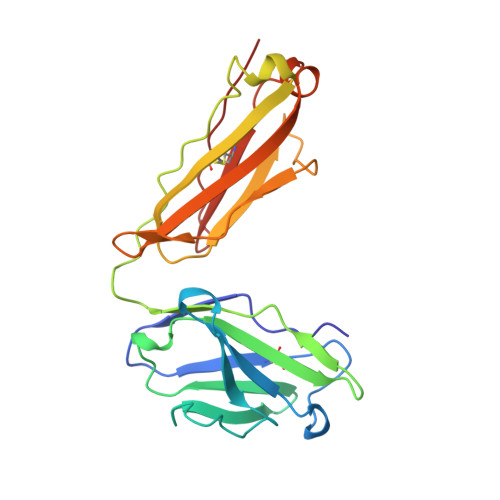
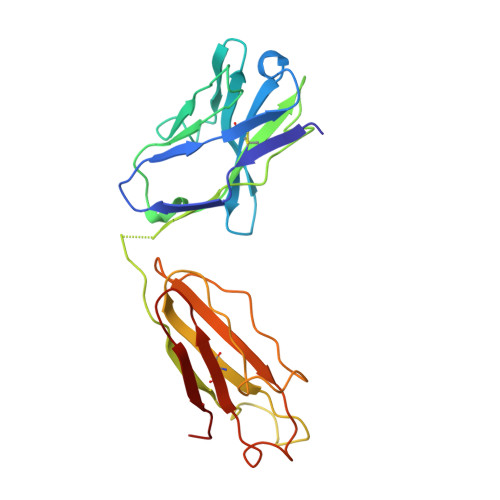
Structural analysis of affinity maturation: the three-dimensional structures of complexes of an anti-nitrophenol antibody.
Yuhasz, S.C., Parry, C., Strand, M., Amzel, L.M.(1995) Mol Immunol 32: 1143-1155
- PubMed: 8544863
- DOI: https://doi.org/10.1016/0161-5890(95)00063-1
- Primary Citation of Related Structures:
1YUH - PubMed Abstract:
Affinity maturation of the immune response to nitrophenol-containing antigens has been extensively investigated. Significant strides made during the past several years with the advent of PCR technology have provided a wealth of biochemical knowledge. Structural investigations of the phenomena have however been limited. We have determined the three-dimensional structure of the Fab fragment of 88C6/12, an anti-4-hydroxy-3-nitrophenyl acetic acid antibody complexed with the immunizing hapten and with a heteroclitic iodinated hapten. The crystallographic structure of the complexes reveals that the binding is stabilized by a number of hydrogen bonds and extensive van der Waals interactions between the hapten and the antibody. In addition, the Fab binding pocket contains a region of positive electrostatic potential well suited for interaction with the predominant resonance form of the nitrophenyl ring system. The observed heteroclicity towards the iodinated hapten is not a direct result of iodine-protein interactions, but results from the enhanced stability in the iodinated ring of the resonance form that binds the antibody. In addition this investigation provides a rationale for the strong preference for the substitution in the heavy chain from the germ-line gene encoded Trp 33 to Leu 33 in the mature anti-nitrophenol response.
Organizational Affiliation:
Department of Biophysics and Biophysical Chemistry, Johns Hopkins School of Medicine, Baltimore, MD 21205, USA.