Crystal Structure of Escherichia coli RNase D, an Exoribonuclease Involved in Structured RNA Processing
Zuo, Y., Wang, Y., Malhotra, A.(2005) Structure 13: 973-984
- PubMed: 16004870
- DOI: https://doi.org/10.1016/j.str.2005.04.015
- Primary Citation of Related Structures:
1YT3 - PubMed Abstract:
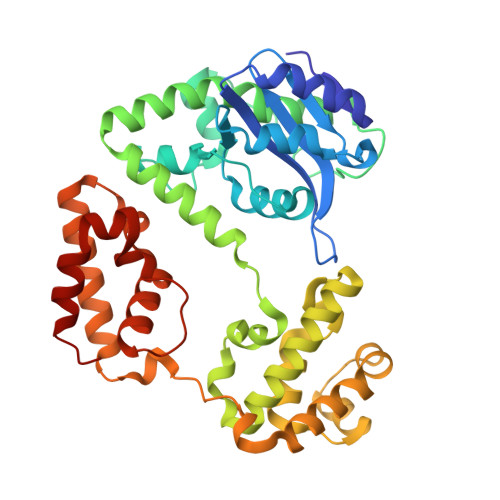
RNase D (RND) is one of seven exoribonucleases identified in Escherichia coli. RNase D has homologs in many eubacteria and eukaryotes, and has been shown to contribute to the 3' maturation of several stable RNAs. Here, we report the 1.6 A resolution crystal structure of E. coli RNase D. The conserved DEDD residues of RNase D fold into an arrangement very similar to the Klenow fragment exonuclease domain. Besides the catalytic domain, RNase D also contains two structurally similar alpha-helical domains with no discernible sequence homology between them. These closely resemble the HRDC domain previously seen in RecQ-family helicases and several other proteins acting on nucleic acids. More interestingly, the DEDD catalytic domain and the two helical domains come together to form a ring-shaped structure. The ring-shaped architecture of E. coli RNase D and the HRDC domains likely play a major role in determining the substrate specificity of this exoribonuclease.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, University of Miami School of Medicine, PO Box 016129, Miami, FL 33101, USA.