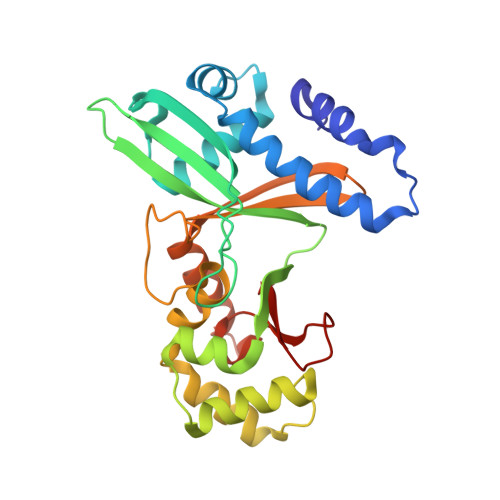
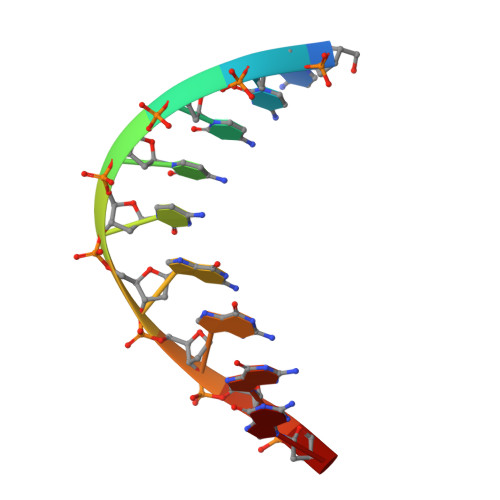
Two crystal forms of the restriction enzyme MspI-DNA complex show the same novel structure.
Xu, Q.S., Roberts, R.J., Guo, H.-C.(2005) Protein Sci 14: 2590-2600
- PubMed: 16195548
- DOI: https://doi.org/10.1110/ps.051565105
- Primary Citation of Related Structures:
1YFI - PubMed Abstract:
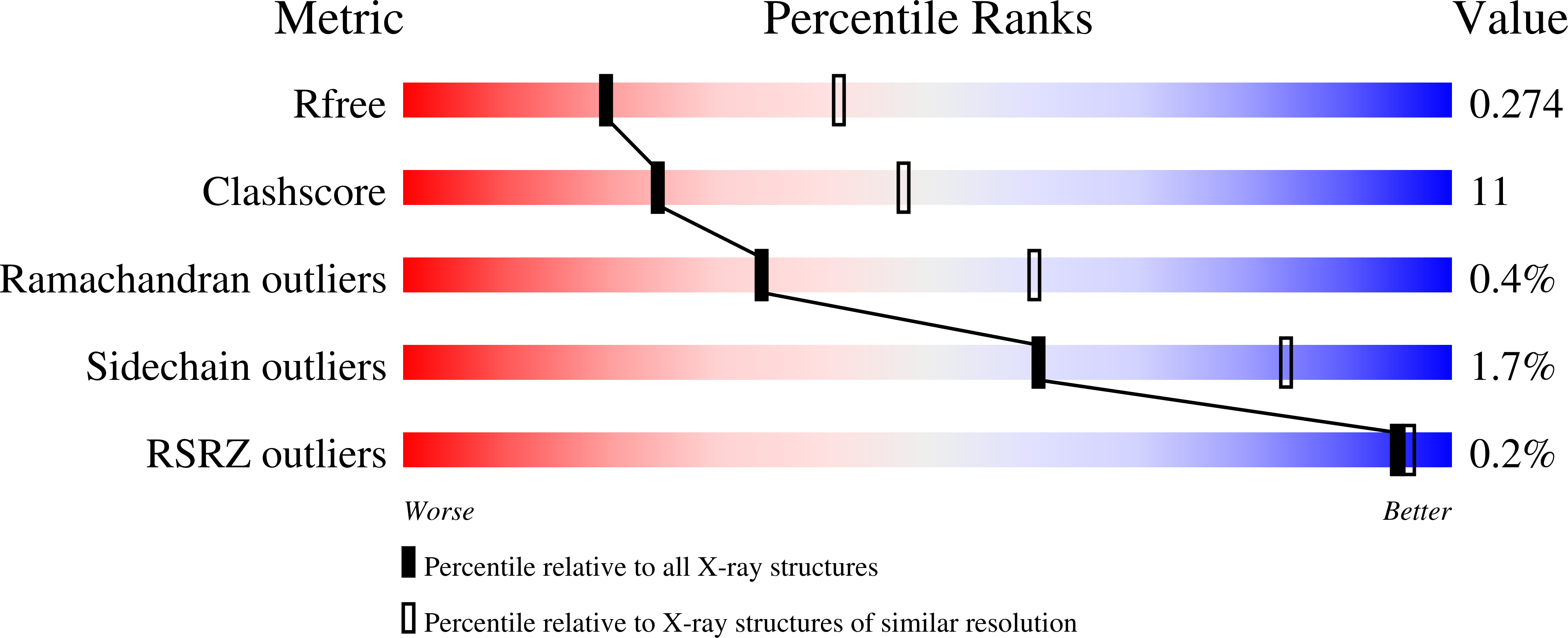
The crystal structure of the Type IIP restriction endonuclease MspI bound to DNA containing its cognate recognition sequence has been determined in both monoclinic and orthorhombic space groups. Significantly, these two independent crystal forms present an identical structure of a novel monomer-DNA complex, suggesting a functional role for this novel enzyme-DNA complex. In both crystals, MspI interacts with the CCGG DNA recognition sequence as a monomer, using an asymmetric mode of recognition by two different structural motifs in a single polypeptide. In the crystallographic asymmetric unit, the two DNA molecules in the two MspI-DNA complexes appear to stack with each other forming an end-to-end pseudo-continuous 19-mer duplex. They are primarily B-form and no major bends or kinks are observed. For DNA recognition, most of the specific contacts between the enzyme and the DNA are preserved in the orthorhombic structure compared with the monoclinic structure. A cation is observed near the catalytic center in the monoclinic structure at a position homologous to the 74/45 metal site of EcoRV, and the orthorhombic structure also shows signs of this same cation. However, the coordination ligands of the metal are somewhat different from those of the 74/45 metal site of EcoRV. Combined with structural information from other solved structures of Type II restriction enzymes, the possible relationship between the structures of the enzymes and their cleavage behaviors is discussed.
Organizational Affiliation:
Department of Physiology and Biophysics, Boston University School of Medicine, Boston, MA 02118, USA. QSXu@lbl.gov