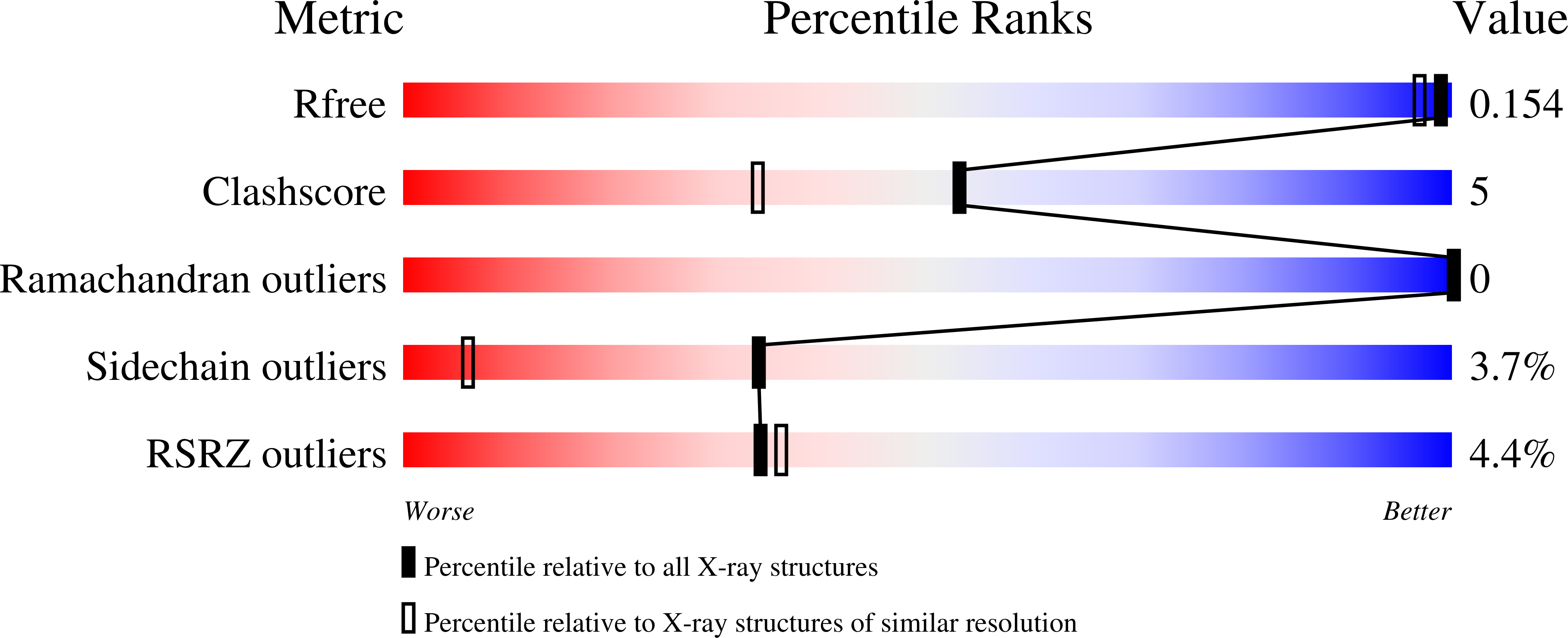
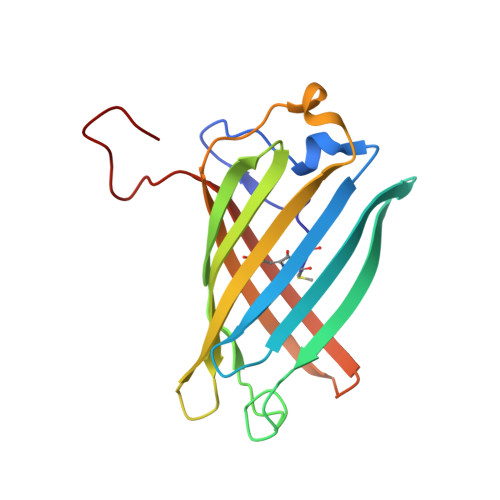
Kindling Fluorescent Protein from Anemonia sulcata: Dark-State Structure at 1.38 Resolution
Quillin, M.L., Anstrom, D.M., Shu, X., O'Leary, S., Kallio, K., Chudakov, D.M., Remington, S.J.(2005) Biochemistry 44: 5774-5787
- PubMed: 15823036
- DOI: https://doi.org/10.1021/bi047644u
- Primary Citation of Related Structures:
1XMZ - PubMed Abstract:
When the nonfluorescent chromoprotein asFP595 from Anemonia sulcata is subjected to sufficiently intense illumination near the absorbance maximum (lambda(abs)(max) = 568 nm), it undergoes a remarkable transition, termed "kindling", to a long-lived fluorescent state (lambda(em)(max) = 595 nm). In the dark recovery phase, the kindled state relaxes thermally on a time scale of seconds or can instantly be reverted upon illumination at 450 nm. The kindling phenomenon is enhanced by the Ala143 --> Gly point mutation, which slows the dark recovery time constant to 100 s at room temperature and increases the fluorescence quantum yield. To investigate the chemical nature of the chromophore and the possible role of chromophore isomerization in the kindling phenomenon, we determined the crystal structure of the "kindling fluorescent protein" asFP595-A143G (KFP) in the dark-adapted state at 1.38 A resolution and 100 K. The chromophore, derived from the Met63-Tyr64-Gly65 tripeptide, closely resembles that of the nonfluorescent chromoprotein Rtms5 in that the configuration is trans about the methylene bridge and there is substantial distortion from planarity. Unlike in Rtms5, in the native protein the polypeptide backbone is cleaved between Cys62 and Met63. The size and shape of the chromophore pocket suggest that the cis isomer of the chromophore could also be accommodated. Within the pocket, partially disordered His197 displays two conformations, which may constitute a binary switch that stabilizes different chromophore configurations. The energy barrier for thermal relaxation was found by Arrhenius plot analysis to be approximately 71 kJ/mol, somewhat higher than the value of approximately 55 kJ/mol observed for cis-trans isomerization of a model chromophore in solution.
Organizational Affiliation:
Department of Physics, Institute of Molecular Biology, University of Oregon, Eugene, Oregon 97403, USA.