Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection
Lei, M., Podell, E.R., Cech, T.R.(2004) Nat Struct Mol Biol 11: 1223-1229
- PubMed: 15558049
- DOI: https://doi.org/10.1038/nsmb867
- Primary Citation of Related Structures:
1XJV - PubMed Abstract:
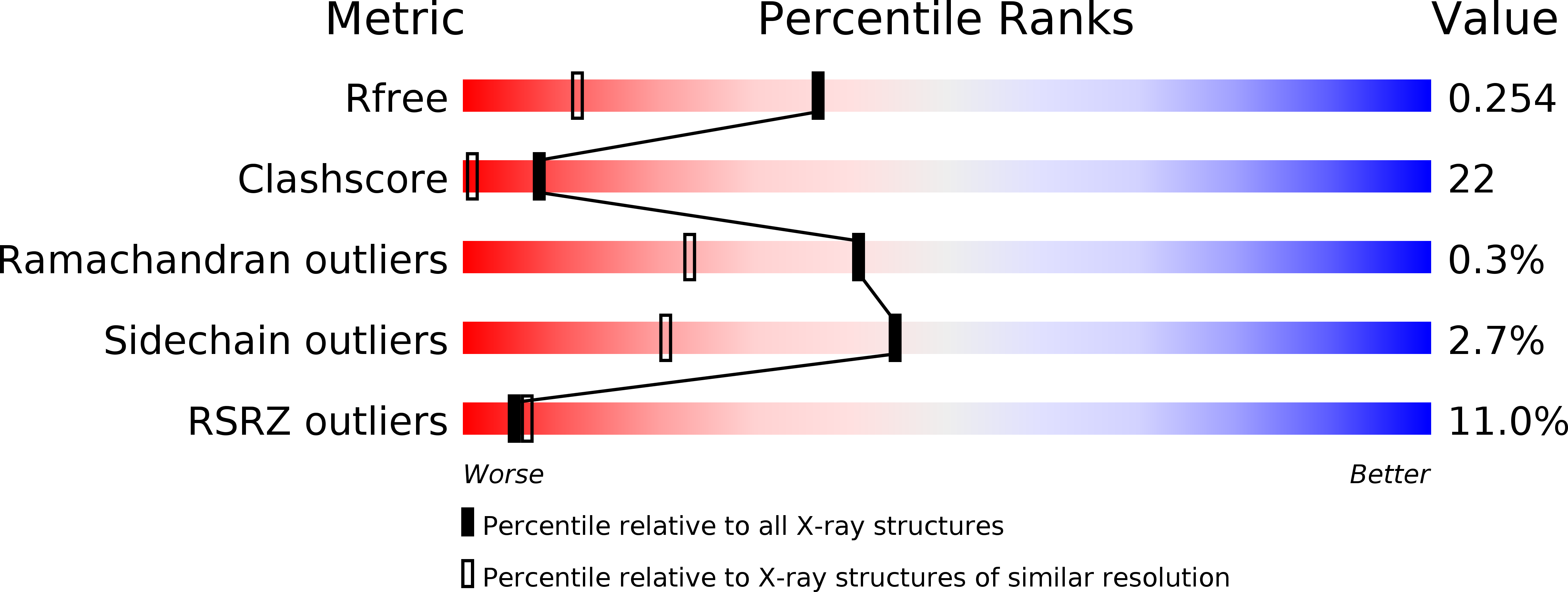
The POT1 (protection of telomeres 1) protein binds the single-stranded overhang at the ends of chromosomes in diverse eukaryotes. It is essential for chromosome end-protection in the fission yeast Schizosaccharomyces pombe, and it is involved in regulation of telomere length in human cells. Here, we report the crystal structure at a resolution of 1.73 A of the N-terminal half of human POT1 (hPOT1) protein bound to a telomeric single-stranded DNA (ssDNA) decamer, TTAGGGTTAG, the minimum tight-binding sequence indicated by in vitro binding assays. The structure reveals that hPOT1 contains two oligonucleotide/ oligosaccharide-binding (OB) folds; the N-terminal OB fold binds the first six nucleotides, resembling the structure of the S. pombe Pot1pN-ssDNA complex, whereas the second OB fold binds and protects the 3' end of the ssDNA. These results provide an atomic-resolution model for chromosome end-capping.
Organizational Affiliation:
Howard Hughes Medical Institute, Department of Chemistry and Biochemistry, University of Colorado, Boulder, Colorado 80309-0215, USA.