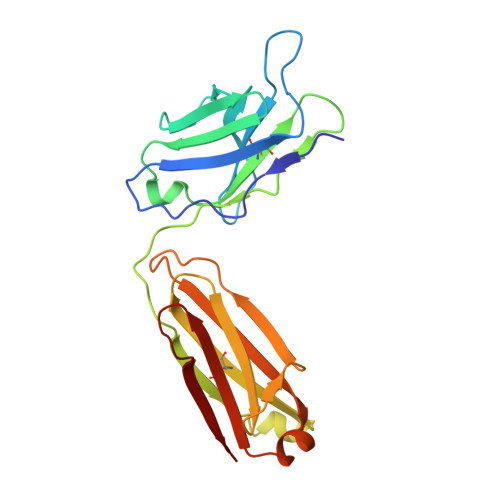
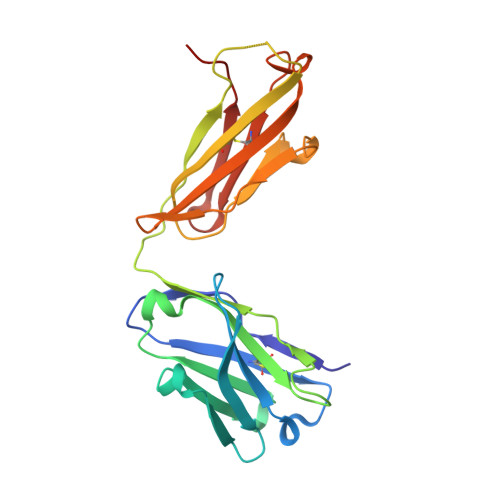
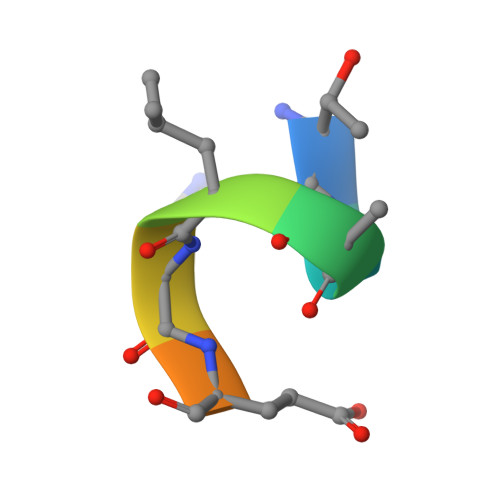
Equilibrium between metarhodopsin-I and metarhodopsin-II is dependent on the conformation of the third cytoplasmic loop.
Piscitelli, C.L., Angel, T.E., Bailey, B.W., Hargrave, P., Dratz, E.A., Lawrence, C.M.(2006) J Biol Chem 281: 6813-6825
- PubMed: 16407202
- DOI: https://doi.org/10.1074/jbc.M510175200
- Primary Citation of Related Structures:
1XGY - PubMed Abstract:
Rhodopsin is a G-protein-coupled receptor (GPCR) that is the light detector in the rod cells of the eye. Rhodopsin is the best understood member of the large GPCR superfamily and is the only GPCR for which atomic resolution structures have been determined. However, these structures are for the inactive, dark-adapted form. Characterization of the conformational changes in rhodopsin caused by light-induced activation is of wide importance, because the metarhodopsin-II photoproduct is analogous to the agonist-occupied conformation of other GPCRs, and metarhodopsin-I may be similar to antagonist-occupied GPCR conformations. In this work we characterize the interaction of antibody K42-41L with the metarhodopsin photoproducts. K42-41L is shown to inhibit formation of metarhodopsin-II while it stabilizes the metarhodopsin-I state. Thus, K42-41L recognizes an epitope accessible in dark-adapted rhodopsin and metarhodopsin-I that is lost upon formation of metarhodopsin-II. Previous work has shown that the peptide TGALQERSK is able to mimic the K42-41L epitope, and we have now determined the structure of the K42-41L-peptide complex. The structure demonstrates a central role for elements of the rhodopsin C3 loop, particularly Gln238 and Glu239, in the interaction with K42-41L. Geometric constraints taken from the antibody-bound peptide were used to model the epitope on the rhodopsin surface. The resulting model suggests that K42-41L locks the C3 loop into an extended conformation that is intermediate between two compact conformations seen in crystal structures of dark-adapted rhodopsin. Together, the structural and functional data strongly suggest that the equilibrium between metarhodopsin-I and metarhodopsin-II is dependent upon the conformation of the C3 loop. The biological implications of this model and its possible relations to dimeric and multimeric complexes of rhodopsin are discussed.
Organizational Affiliation:
Department of Chemistry, Montana State University, Bozeman, Montana 59717, USA.