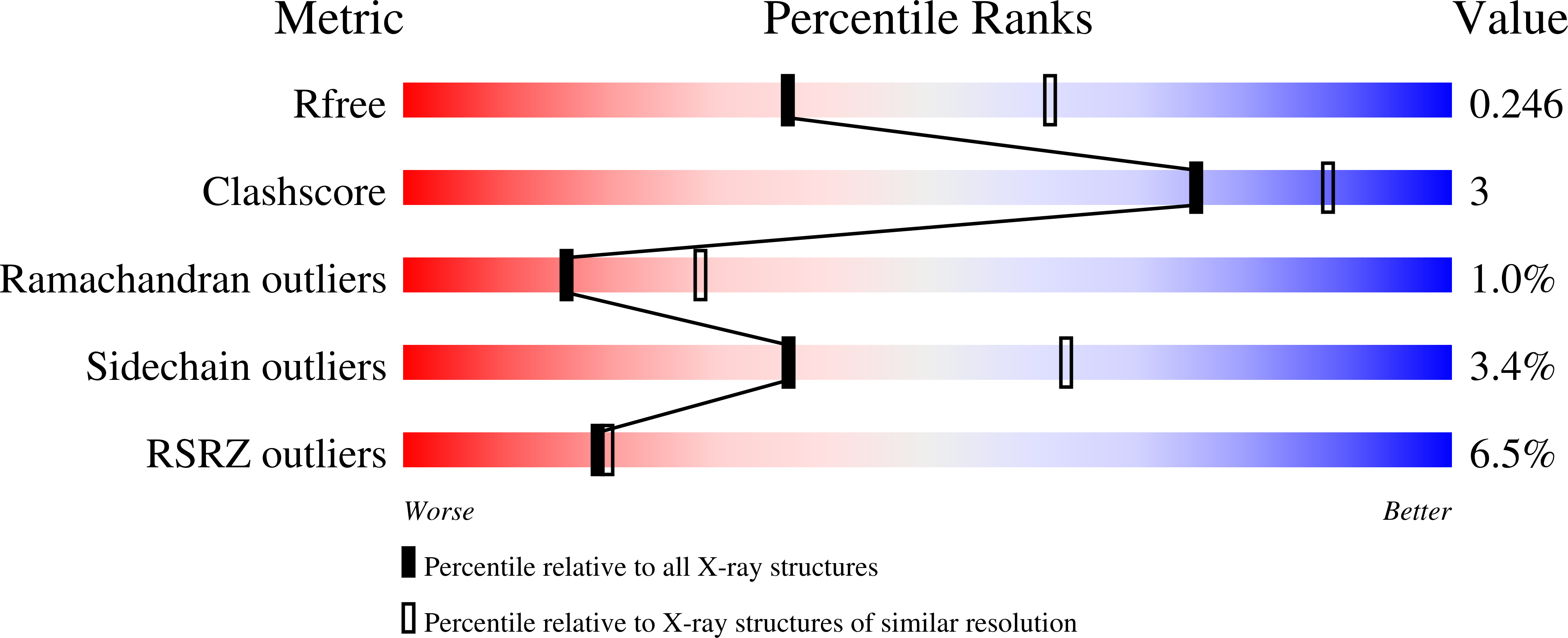
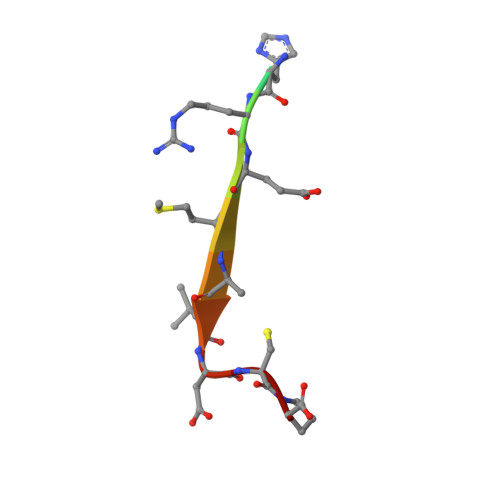
Internal recognition through PDZ domain plasticity in the Par-6-Pals1 complex.
Penkert, R.R., Divittorio, H.M., Prehoda, K.E.(2004) Nat Struct Mol Biol 11: 1122-1127
- PubMed: 15475968
- DOI: https://doi.org/10.1038/nsmb839
- Primary Citation of Related Structures:
1X8S - PubMed Abstract:
PDZ protein interaction domains are typically selective for C-terminal ligands, but non-C-terminal, 'internal' ligands have also been identified. The PDZ domain from the cell polarity protein Par-6 binds C-terminal ligands and an internal sequence from the protein Pals1/Stardust. The structure of the Pals1-Par-6 PDZ complex reveals that the PDZ ligand-binding site is deformed to allow for internal binding. Whereas binding of the Rho GTPase Cdc42 to a CRIB domain adjacent to the Par-6 PDZ regulates binding of C-terminal ligands, the conformational change that occurs upon binding of Pals1 renders its binding independent of Cdc42. These results suggest a mechanism by which the requirement for a C terminus can be readily bypassed by PDZ ligands and reveal a complex set of cooperative and competitive interactions in Par-6 that are likely to be important for cell polarity regulation.
Organizational Affiliation:
Institute of Molecular Biology and Department of Chemistry, University of Oregon, Eugene, Oregon 97403, USA.