Structural basis for the assembly of a nuclear export complex.
Matsuura, Y., Stewart, M.(2004) Nature 432: 872-877
Experimental Data Snapshot
(2004) Nature 432: 872-877
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| GTP-binding nuclear protein Ran | 176 | Canis lupus familiaris | Mutation(s): 0 Gene Names: RAN | 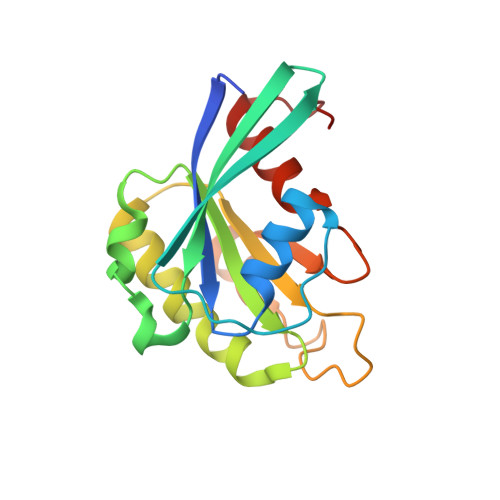 | |
UniProt | |||||
Find proteins for P62825 (Canis lupus familiaris) Explore P62825 Go to UniProtKB: P62825 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P62825 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| SRP1 isoform 1 | 530 | Saccharomyces cerevisiae | Mutation(s): 0 Gene Names: SRP1, GI526_G0004888 | 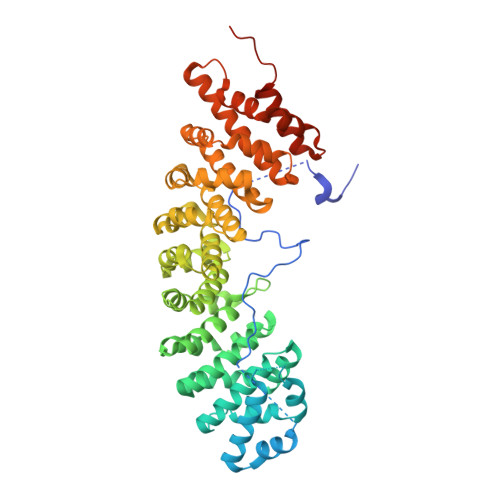 | |
UniProt | |||||
Find proteins for Q02821 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore Q02821 Go to UniProtKB: Q02821 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q02821 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Importin alpha re-exporter | 960 | Saccharomyces cerevisiae | Mutation(s): 0 Gene Names: CSE1, YGL238W, HRC135 | 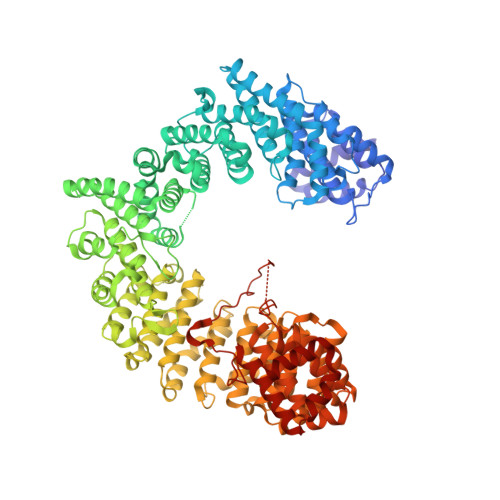 | |
UniProt | |||||
Find proteins for P33307 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore P33307 Go to UniProtKB: P33307 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P33307 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| GTP Query on GTP | D [auth A] | GUANOSINE-5'-TRIPHOSPHATE C10 H16 N5 O14 P3 XKMLYUALXHKNFT-UUOKFMHZSA-N |  | ||
| MG Query on MG | E [auth A] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 272.672 | α = 90 |
| b = 72.261 | β = 92.57 |
| c = 97.64 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| MOSFLM | data reduction |
| SCALA | data scaling |
| CNS | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Human Frontier Science Program (HFSP) | -- |