The Structure of Cyclin E1/Cdk2: Implications for Cdk2 Activation and Cdk2-Independent Roles
Honda, R., Lowe, E.D., Dubinina, E., Skamnaki, V., Cook, A., Brown, N., Johnson, L.N.(2005) EMBO J 24: 452
- PubMed: 15660127
- DOI: https://doi.org/10.1038/sj.emboj.7600554
- Primary Citation of Related Structures:
1W98 - PubMed Abstract:
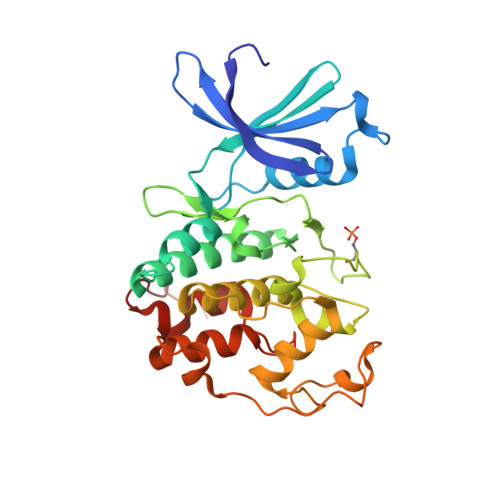
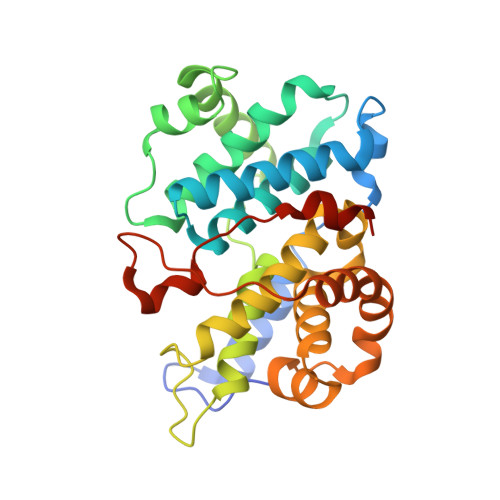
Cyclin E, an activator of phospho-CDK2 (pCDK2), is important for cell cycle progression in metazoans and is frequently overexpressed in cancer cells. It is essential for entry to the cell cycle from G0 quiescent phase, for the assembly of prereplication complexes and for endoreduplication in megakaryotes and giant trophoblast cells. We report the crystal structure of pCDK2 in complex with a truncated cyclin E1 (residues 81-363) at 2.25 A resolution. The N-terminal cyclin box fold of cyclin E1 is similar to that of cyclin A and promotes identical changes in pCDK2 that lead to kinase activation. The C-terminal cyclin box fold shows significant differences from cyclin A. It makes additional interactions with pCDK2, especially in the region of the activation segment, and contributes to CDK2-independent binding sites of cyclin E. Kinetic analysis with model peptide substrates show a 1.6-fold increase in kcat for pCDK2/cyclin E1 (81-363) over kcat of pCDK2/cyclin E (full length) and pCDK2/cyclin A. The structural and kinetic results indicate no inherent substrate discrimination between pCDK2/cyclin E and pCDK2/cyclin A with model substrates.
Organizational Affiliation:
Laboratory of Molecular Biophysics, Department of Biochemistry, University of Oxford, Oxford, UK.