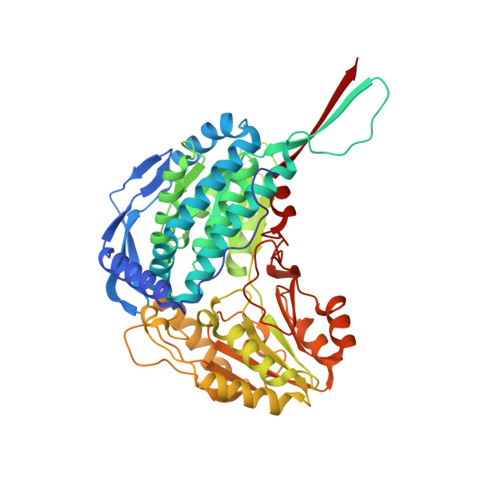
Crystal Structure of Thermus Thermophilus Delta(1)- Pyrroline-5-Carboxylate Dehydrogenase.
Inagaki, E., Ohshima, N., Takahashi, H., Kuroishi, C., Yokoyama, S., Tahirov, T.H.(2006) J Mol Biol 362: 490
- PubMed: 16934832
- DOI: https://doi.org/10.1016/j.jmb.2006.07.048
- Primary Citation of Related Structures:
1UZB, 2BHP, 2BHQ, 2BJA, 2BJK, 2IY6 - PubMed Abstract:
Delta(1)-pyrroline-5-carboxylate dehydrogenase (P5CDh) plays an important role in the metabolic pathway from proline to glutamate. It irreversibly catalyzes the oxidation of glutamate-gamma-semialdehyde, the product of the non-enzymatic hydrolysis of Delta(1)-pyrroline-5-carboxylate, into glutamate with the reduction of NAD(+) into NADH. We have confirmed the P5CDh activity of the Thermus thermophilus protein TT0033 (TtP5CDh), and determined the crystal structure of the enzyme in the ligand-free form at 1.4 A resolution. To investigate the structural basis of TtP5CDh function, the TtP5CDh structures with NAD(+), with NADH, and with its product glutamate were determined at 1.8 A, 1.9 A, and 1.4 A resolution, respectively. The solved structures suggest an overall view of the P5CDh catalytic mechanism and provide insights into the P5CDh deficiencies in the case of the human type II hyperprolinemia.
Organizational Affiliation:
RIKEN SPring-8 Center, Harima Institute, 1-1-1 Kouto, Sayo, Hyogo 679-5148, Japan.