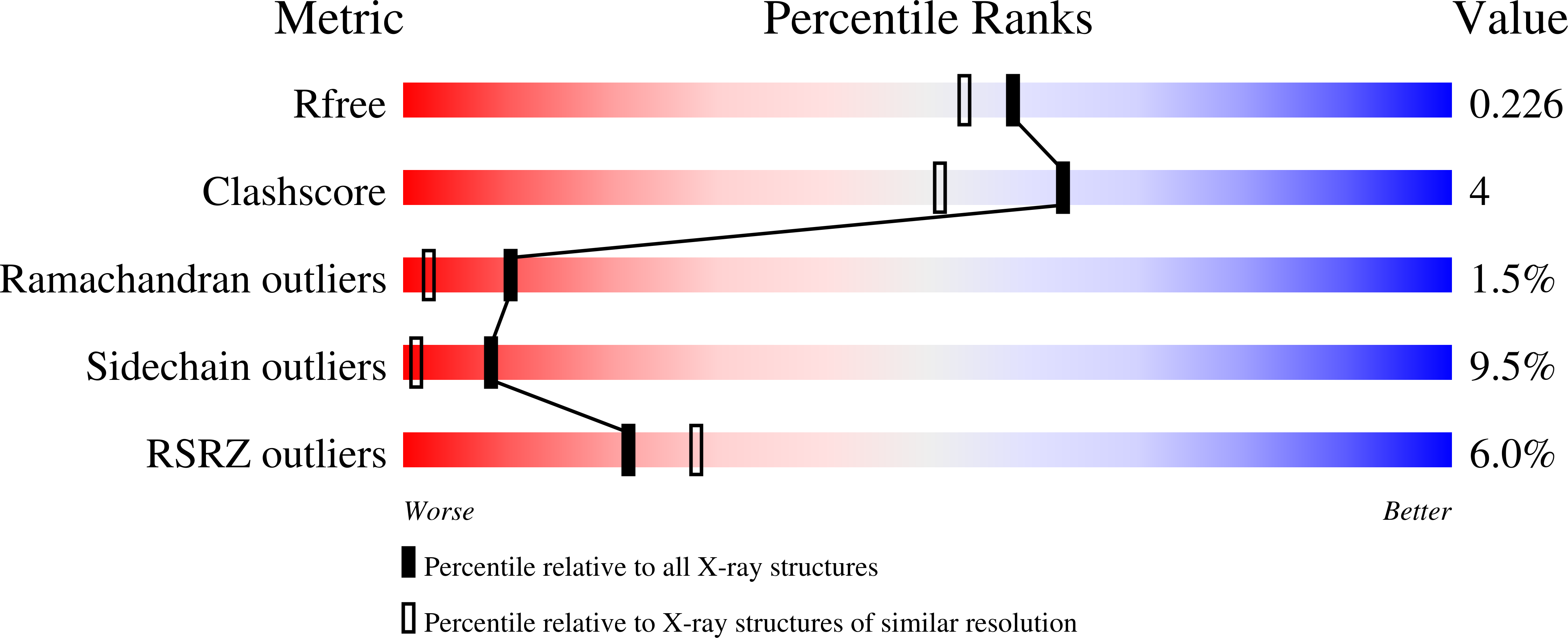
A Novel Adp- and Zinc-Binding Fold from Function-Directed in Vitro Evolution
Lo Surdo, P., Walsh, M.A., Sollazzo, M.(2004) Nat Struct Mol Biol 11: 382
- PubMed: 15024384
- DOI: https://doi.org/10.1038/nsmb745
- Primary Citation of Related Structures:
1UW1 - PubMed Abstract:
A great challenge to biologists is to create proteins with novel folds and tailored functions. As an alternative to de novo protein design, we investigated the structure of a randomly generated protein targeted to bind ATP. The crystal structure reveals a novel alpha/beta fold bound to its ligand, representing both the first protein structure derived from in vitro evolution and the first nucleotide-binding protein stabilized by a zinc ion.
Organizational Affiliation:
Istituto di Ricerche di Biologia Molecolare (IRBM), Via Pontina Km 30.600, 00040 Pomezia, Roma, Italy. paola_losurdo@merck.com