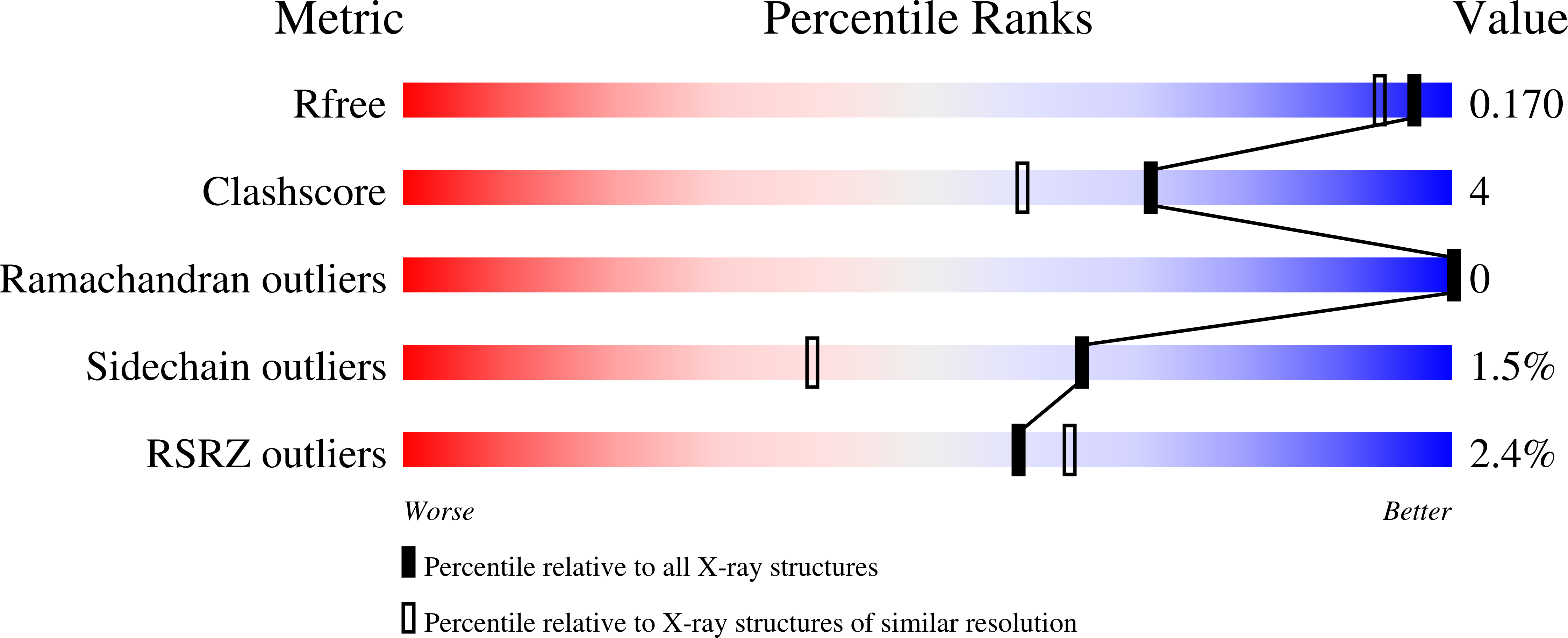
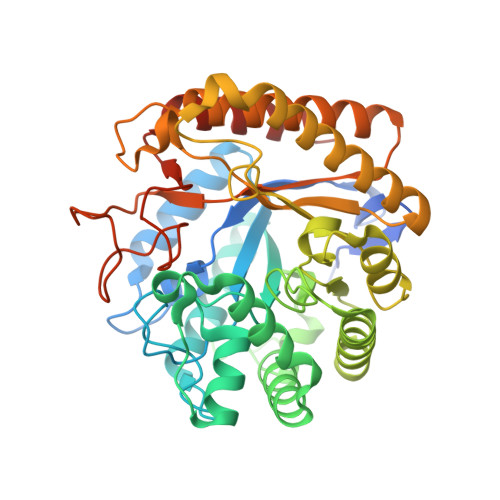
Insights Into the Molecular Determinants of Substrate Specificity in Glycoside Hydrolase Family 5 Revealed by the Crystal Structure and Kinetics of Cellvibrio Mixtus Mannosidase 5A
Dias, M.V.F., Vincent, F., Pell, G., Prates, J.A.M., Centeno, M.S.J., Ferreira, L.M.A., Gilbert, H.J., Davies, G.J., Fontes, C.M.G.A.(2004) J Biol Chem 279: 25517
- PubMed: 15014076
- DOI: https://doi.org/10.1074/jbc.M401647200
- Primary Citation of Related Structures:
1UUQ - PubMed Abstract:
The enzymatic hydrolysis of the glycosidic bond is central to numerous biological processes. Glycoside hydrolases, which catalyze these reactions, are grouped into families based on primary sequence similarities. One of the largest glycoside hydrolase families is glycoside hydrolase family 5 (GH5), which contains primarily endo-acting enzymes that hydrolyze beta-mannans and beta-glucans. Here we report the cloning, characterization, and three-dimensional structure of the Cellvibrio mixtus GH5 beta-mannosidase (CmMan5A). This enzyme releases mannose from the nonreducing end of mannooligosaccharides and polysaccharides, an activity not previously observed in this enzyme family. CmMan5A contains a single glycone (-1) and two aglycone (+1 and +2) sugar-binding subsites. The -1 subsite displays absolute specificity for mannose, whereas the +1 subsite does not accommodate galactosyl side chains but will bind weakly to glucose. The +2 subsite is able to bind to decorated mannose residues. CmMan5A displays similar activity against crystalline and amorphous mannans, a property rarely attributed to glycoside hydrolases. The 1.5 A crystal structure reveals that CmMan5A adopts a (beta/alpha)(8) barrel fold, and superimposition with GH5 endo-mannanases shows that dramatic differences in the length of three loops modify the active center accessibility and thus modulate the specificity from endo to exo. The most striking and significant difference is the extended loop between strand beta8 and helix alpha8 comprising residues 378-412. This insertion forms a "double" steric barrier, formed by two short beta-strands that function to "block" the substrate binding cleft at the edge of the -1 subsite forming the "exo" active center topology of CmMan5A.
Organizational Affiliation:
CIISA-Faculdade de Medicina Veterinária, Universidade Técnica de Lisboa, Rua Prof. Cid dos Santos, 1300-477 Lisboa, Portugal.