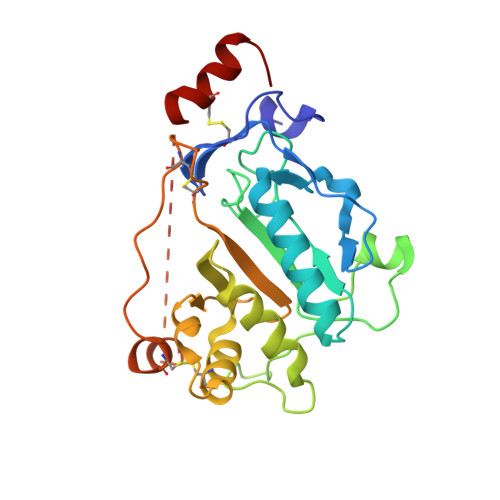
Peptidoglycan amidase MepA is a LAS metallopeptidase
Marcyjaniak, M., Odintsov, S.G., Sabala, I., Bochtler, M.(2004) J Biol Chem 279: 43982-43989
- PubMed: 15292190
- DOI: https://doi.org/10.1074/jbc.M406735200
- Primary Citation of Related Structures:
1TZP, 1U10 - PubMed Abstract:
LAS enzymes are a group of metallopeptidases that share an active site architecture and a core folding motif and have been named according to the group members lysostaphin, D-Ala-D-Ala carboxypeptidase and sonic hedgehog. Escherichia coli MepA is a periplasmic, penicillin-insensitive murein endopeptidase that cleaves the D-alanyl-meso-2,6-diamino-pimelyl amide bond in E. coli peptidoglycan. The enzyme lacks sequence similarity with other peptidases, and is currently classified as a peptidase of unknown fold and catalytic class in all major data bases. Here, we build on our observation that two motifs, characteristic of the newly described LAS group of metallopeptidases, are conserved in MepA-type sequences. We demonstrate that recombinant E. coli MepA is sensitive to metal chelators and that mutations in the predicted Zn2+ ligands His-113, Asp-120, and His-211 inactivate the enzyme. Moreover, we present the crystal structure of MepA. The active site of the enzyme is most similar to the active sites of lysostaphin and D-Ala-D-Ala carboxypeptidase, and the fold is most closely related to the N-domain of sonic hedgehog. We conclude that MepA-type peptidases are LAS enzymes.
Organizational Affiliation:
International Institute of Molecular and Cell Biology, ul. Trojdena 4, 02-109 Warsaw, Poland.