CRYSTAL STRUCTURE OF YEAST HYPOTHETICAL PROTEIN YMX7
Kumaran, D., Swaminathan, S.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Hypothetical 32.1 kDa protein in ADH3-RCA1 intergenic region | 284 | Saccharomyces cerevisiae | Mutation(s): 0 Gene Names: YMR087W, YM9582.12 | 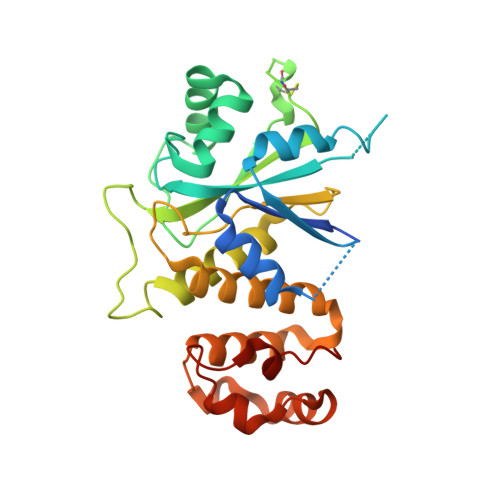 | |
UniProt | |||||
Find proteins for Q04299 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore Q04299 Go to UniProtKB: Q04299 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q04299 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ADP Query on ADP | E [auth A] | ADENOSINE-5'-DIPHOSPHATE C10 H15 N5 O10 P2 XTWYTFMLZFPYCI-KQYNXXCUSA-N |  | ||
| CL Query on CL | D [auth A] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| NA Query on NA | B [auth A], C [auth A] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 107.948 | α = 90 |
| b = 38.054 | β = 111.75 |
| c = 63.137 | γ = 90 |
| Software Name | Purpose |
|---|---|
| CBASS | data collection |
| HKL-2000 | data reduction |
| CNS | refinement |
| HKL-2000 | data scaling |