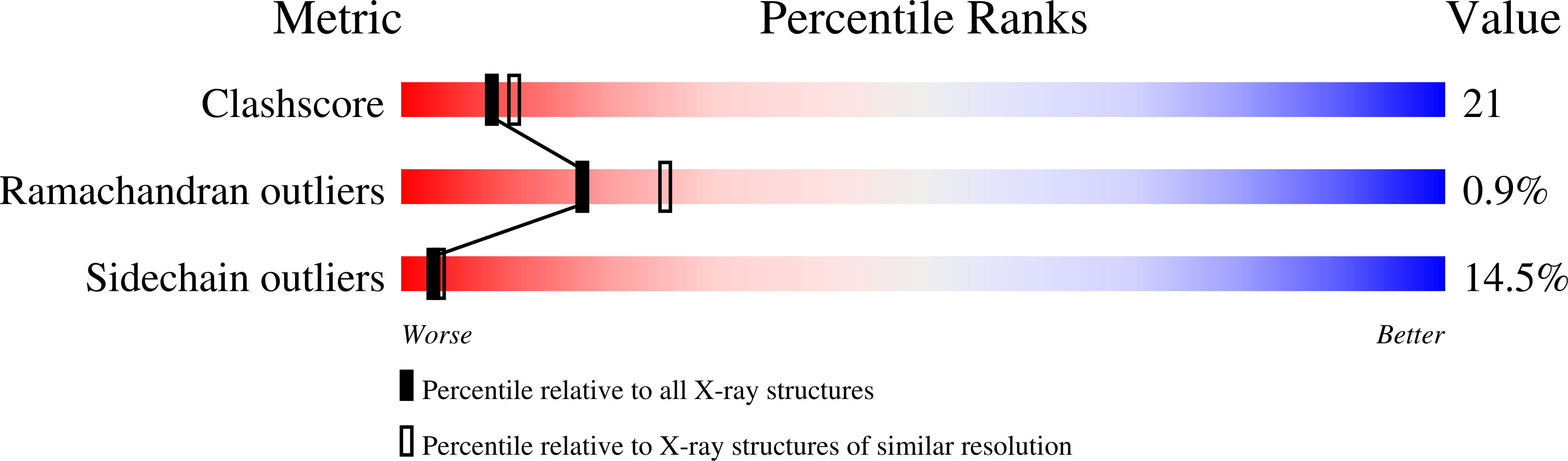The crystal structure of an engineered monomeric triosephosphate isomerase, monoTIM: the correct modelling of an eight-residue loop.
Borchert, T.V., Abagyan, R., Kishan, K.V., Zeelen, J.P., Wierenga, R.K.(1993) Structure 1: 205-213
- PubMed: 16100954
- DOI: https://doi.org/10.1016/0969-2126(93)90021-8
- Primary Citation of Related Structures:
1TRI - PubMed Abstract:
The triosephosphate isomerase (TIM) fold is found in several different classes of enzymes, most of which are oligomers; TIM itself always functions as a very tight dimer. It has recently been shown that a monomeric form of TIM ('monoTIM') can be constructed by replacing a 15-residue interface loop, loop-3, with an eight-residue fragment; modelling suggests that this should result in a short strain-free turn, resulting in the subsequent helix, helix-A3, having an additional turn at its amino terminus.
Organizational Affiliation:
European Molecular Biology Laboratory, Meyerhofstrasse 1, Postfach 102209, D69012 Heidelberg, Germany.