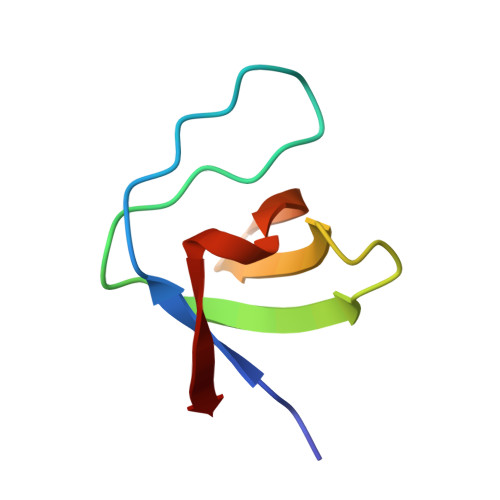
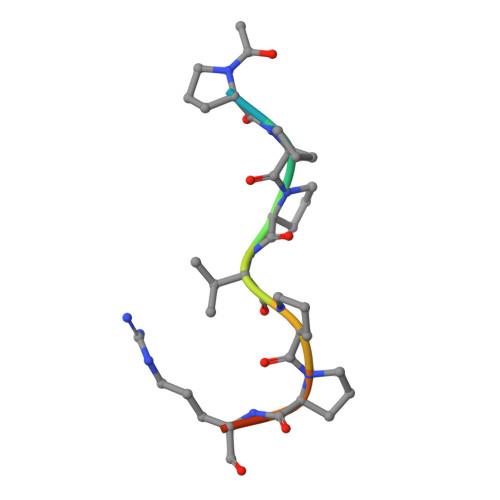
Structural determinants of peptide-binding orientation and of sequence specificity in SH3 domains.
Lim, W.A., Richards, F.M., Fox, R.O.(1994) Nature 372: 375-379
- PubMed: 7802869
- DOI: https://doi.org/10.1038/372375a0
- Primary Citation of Related Structures:
1SEM - PubMed Abstract:
The Src-homology-3 (SH3) domains of the Caenorhabditis elegans protein SEM-5 and its human and Drosophila homologues, Grb2 and Drk (refs 1-4), bind proline-rich sequences found in the nucleotide-exchange factor Sos as part of their proposed function linking receptor tyrosine kinase activation to Ras activation. Here we report the crystal structure at 2.0 A resolution of the carboxy-terminal SH3 domain from SEM-5 complexed to the mSos-derived amino-acid sequence PPPVPPRRR. The peptide is found to bind in an orientation ('minus') that is precisely opposite to that observed previously ('plus' orientation) in other SH3-peptide complexes. This novel ability of peptide-recognition proteins to recognize peptides in two distinct modes may play an important role in the signalling specificity of pathways involving SH3 domains. Comparison of this structure with other SH3 complexes reveals how a conserved binding face can be used to recognize peptides in different orientations, and why the Sos peptide binds in this particular orientation.
Organizational Affiliation:
Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, Connecticut 06520-8114.