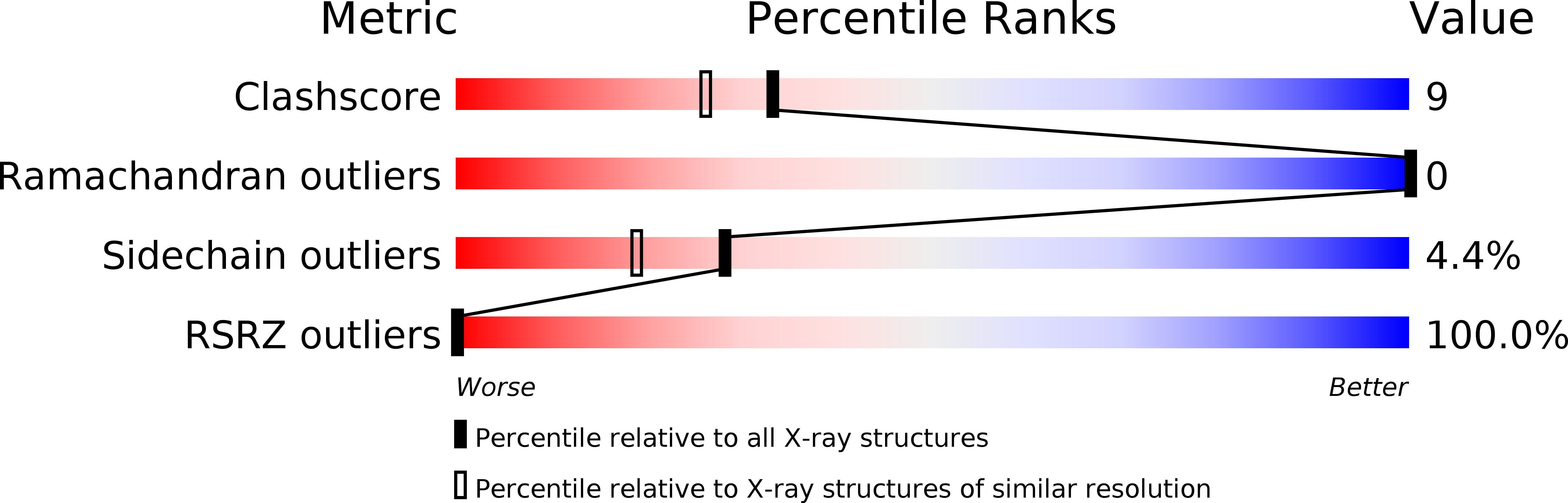
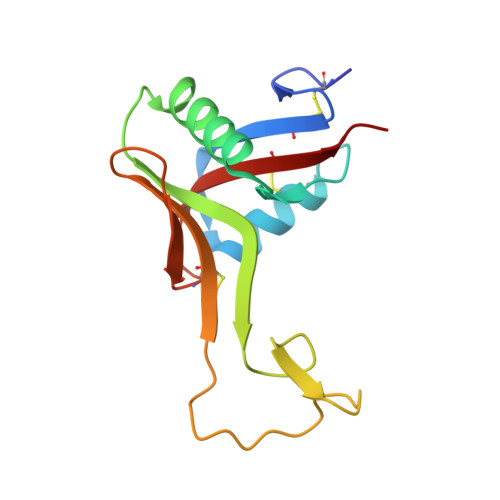
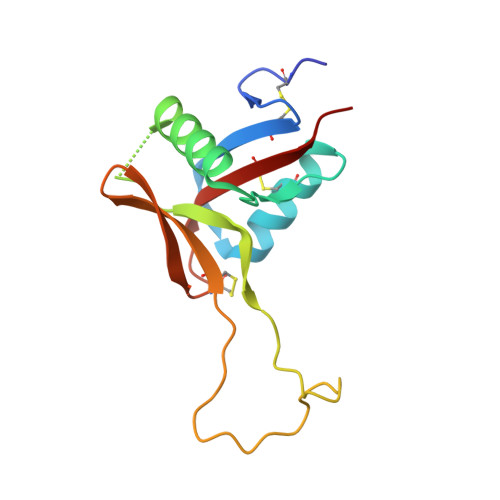
Structure of rhodocetin reveals noncovalently bound heterodimer interface
Paaventhan, P., Kong, C.G., Joseph, J.S., Chung, M.C.M., Kolatkar, P.R.(2005) Protein Sci 14: 169-175
- PubMed: 15576563
- DOI: https://doi.org/10.1110/ps.04945605
- Primary Citation of Related Structures:
1SB2 - PubMed Abstract:
Rhodocetin is a unique heterodimer consisting of alpha- and beta-subunits of 133 and 129 residues, respectively. The molecule, purified from the crude venom of the Malayan pit viper, Calloselasma rhodostoma, functions as an inhibitor of collagen-induced aggregation. Rhodocetin has been shown to have activity only when present as a dimer. The dimer is formed without an intersubunit disulfide bridge, unlike all the other Ca(2+)-dependent lectin-like proteins. We report here the 1.9 A resolution structure of rhodocetin, which reveals the compensatory interactions that occur in the absence of the disulfide bridge to preserve activity.
Organizational Affiliation:
Genome Institute of Singapore, Singapore 138672.