The X-ray structure of trans-3-chloroacrylic acid dehalogenase reveals a novel hydration mechanism in the tautomerase superfamily
de Jong, R.M., Brugman, W., Poelarends, G.J., Whitman, C.P., Dijkstra, B.W.(2004) J Biol Chem 279: 11546-11552
- PubMed: 14701869
- DOI: https://doi.org/10.1074/jbc.M311966200
- Primary Citation of Related Structures:
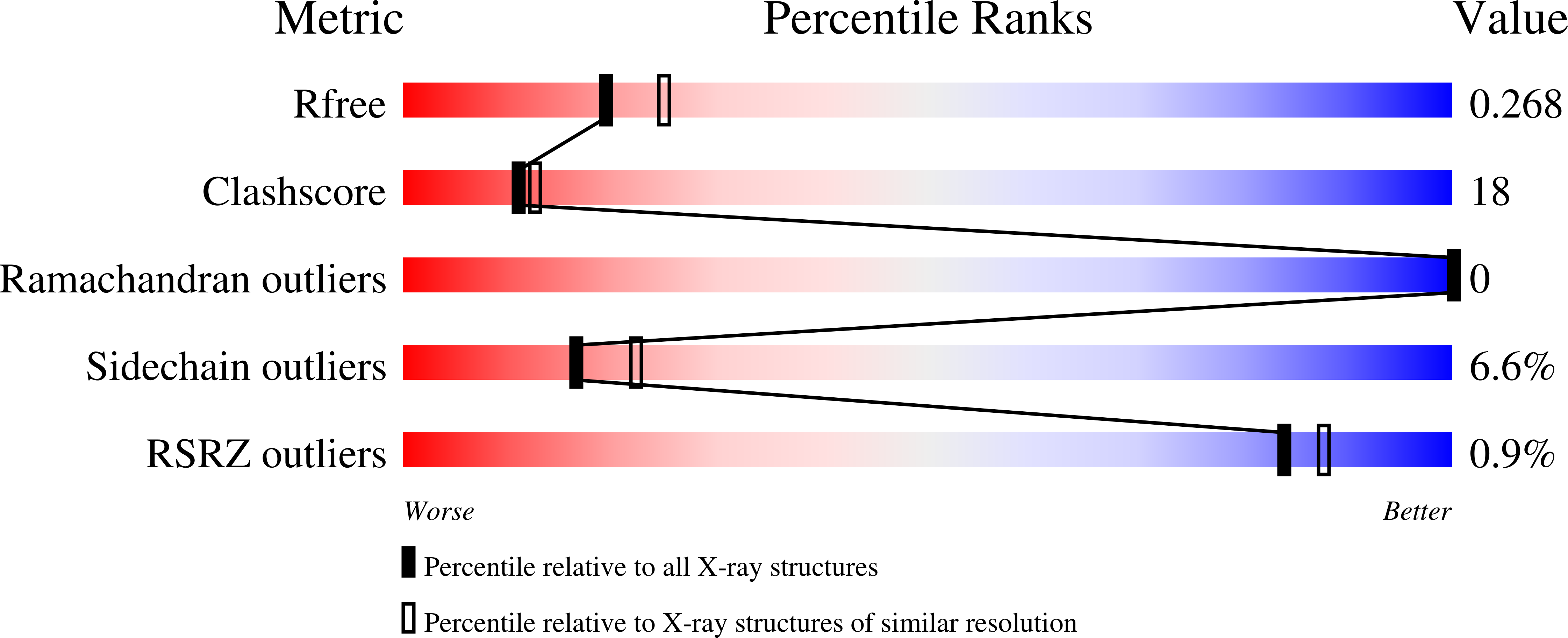
1S0Y - PubMed Abstract:
Isomer-specific 3-chloroacrylic acid dehalogenases function in the bacterial degradation of 1,3-dichloropropene, a compound used in agriculture to kill plant-parasitic nematodes. The crystal structure of the heterohexameric trans-3-chloroacrylic acid dehalogenase (CaaD) from Pseudomonas pavonaceae 170 inactivated by 3-bromopropiolate shows that Glu-52 in the alpha-subunit is positioned to function as the water-activating base for the addition of a hydroxyl group to C-3 of 3-chloroacrylate and 3-bromopropiolate, whereas the nearby Pro-1 in the beta-subunit is positioned to provide a proton to C-2. Two arginine residues, alphaArg-8 and alphaArg-11, interact with the C-1 carboxylate groups, thereby polarizing the alpha,beta-unsaturated acids. The reaction with 3-chloroacrylate results in the production of an unstable halohydrin, 3-chloro-3-hydroxypropanoate, which decomposes into the products malonate semialdehyde and HCl. In the inactivation mechanism, however, malonyl bromide is produced, which irreversibly alkylates the betaPro-1. CaaD is related to 4-oxalocrotonate tautomerase, with which it shares an N-terminal proline. However, in 4-oxalocrotonate tautomerase, Pro-1 functions as a base participating in proton transfer within a hydrophobic active site, whereas in CaaD, the acidic proline is stabilized in a hydrophilic active site. The altered active site environment of CaaD thus facilitates a previously unknown reaction in the tautomerase superfamily, the hydration of the alpha,beta-unsaturated bonds of trans-3-chloroacrylate and 3-bromopropiolate. The mechanism for these hydration reactions represents a novel catalytic strategy that results in carbon-halogen bond cleavage.
Organizational Affiliation:
Laboratory of Biophysical Chemistry, University of Groningen, Nijenborgh 4, 9747 AG Groningen, The Netherlands.