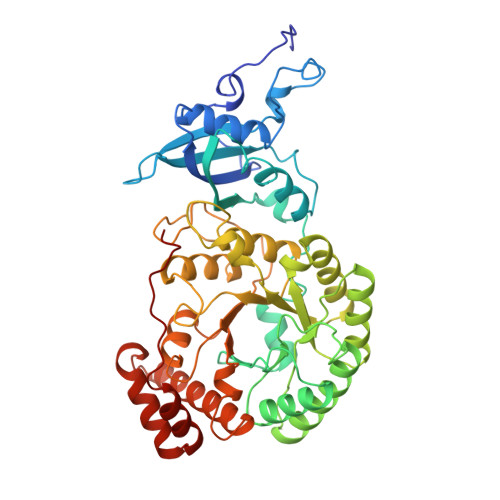
Structure of an effector-induced inactivated state of ribulose 1,5-bisphosphate carboxylase/oxygenase: the binary complex between enzyme and xylulose 1,5-bisphosphate.
Newman, J., Gutteridge, S.(1994) Structure 2: 495-502
- PubMed: 7922027
- DOI: https://doi.org/10.1016/s0969-2126(00)00050-2
- Primary Citation of Related Structures:
1RSC - PubMed Abstract:
Ribulose 1,5-bisphosphate carboxylase/oxygenase (rubisco) catalyzes the addition of CO2 to ribulose 1,5-bisphosphate in all photosynthetic organisms. During catalysis, the bisphosphate is depleted by reactions other than carboxylation and some of the products are potent inhibitors of rubisco. We have used one of these, xylulose 1,5-bisphosphate as an analogue of the natural substrate and co-crystallized it with the enzyme.
Organizational Affiliation:
Department of Molecular Biology, Biomedical Centre, Uppsala, Sweden.