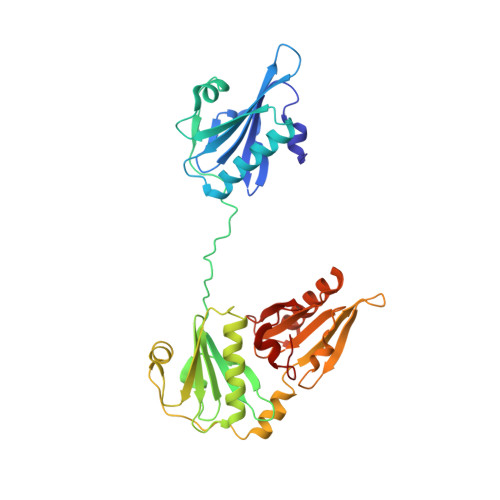
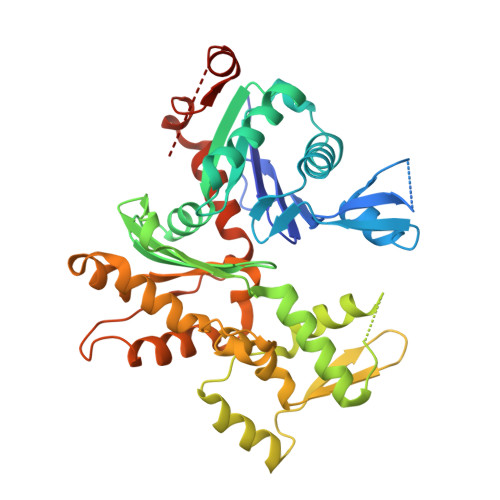
Structure of the N-terminal half of gelsolin bound to actin: roles in severing, apoptosis and FAF
Burtnick, L.D., Urosev, D., Irobi, E., Narayan, K., Robinson, R.C.(2004) EMBO J 23: 2713-2722
- PubMed: 15215896
- DOI: https://doi.org/10.1038/sj.emboj.7600280
- Primary Citation of Related Structures:
1RGI - PubMed Abstract:
The actin filament-severing functionality of gelsolin resides in its N-terminal three domains (G1-G3). We have determined the structure of this fragment in complex with an actin monomer. The structure reveals the dramatic domain rearrangements that activate G1-G3, which include the replacement of interdomain interactions observed in the inactive, calcium-free protein by new contacts to actin, and by a novel G2-G3 interface. Together, these conformational changes are critical for actin filament severing, and we suggest that their absence leads to the disease Finnish-type familial amyloidosis. Furthermore, we propose that association with actin drives the calcium-independent activation of isolated G1-G3 during apoptosis, and that a similar mechanism operates to activate native gelsolin at micromolar levels of calcium. This is the first structure of a filament-binding protein bound to actin and it sets stringent, high-resolution limitations on the arrangement of actin protomers within the filament.
Organizational Affiliation:
Department of Chemistry and Centre for Blood Research, The University of British Columbia, Vancouver, BC, Canada.