NMR Structure of an Anti-Gp120 Antibody Complex with a V3 Peptide Reveals a Surface Important for Co-Receptor Binding
Tugarinov, V., Zvi, A., Levy, R., Hayek, Y., Matsushita, S., Anglister, J.(2000) Structure 8: 385-395
- PubMed: 10801487
- DOI: https://doi.org/10.1016/s0969-2126(00)00119-2
- Primary Citation of Related Structures:
1QNZ - PubMed Abstract:
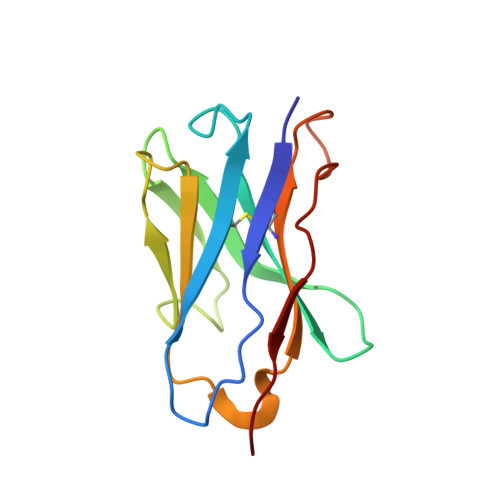
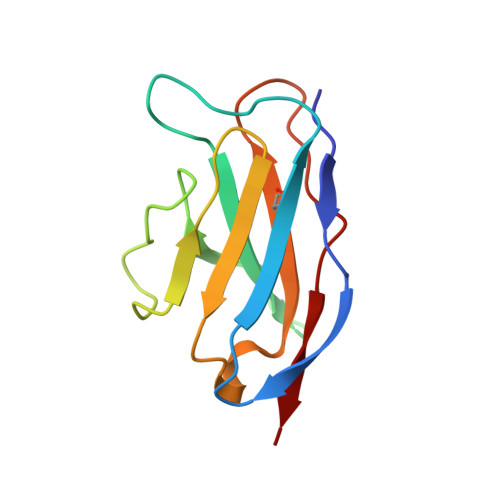
The protein 0.5beta is a potent strain-specific human immunodeficiency virus type 1 (HIV-1) neutralizing antibody raised against the entire envelope glycoprotein (gp120) of the HIV-1(IIIB) strain. The epitope recognized by 0.5beta is located within the third hypervariable region (V3) of gp120. Recently, several HIV-1 V3 residues involved in co-receptor utilization and selection were identified. Virtually complete sidechain assignment of the variable fragment (Fv) of 0.5beta in complex with the V3(IIIB) peptide P1053 (RKSIRIQRGPGRAFVTIG, in single-letter amino acid code) was accomplished and the combining site structure of 0.5beta Fv complexed with P1053 was solved using multidimensional nuclear magnetic resonance (NMR). Five of the six complementarity determining regions (CDRs) of the antibody adopt standard canonical conformations, whereas CDR3 of the heavy chain assumes an unexpected fold. The epitope recognized by 0.5beta encompasses 14 of the 18 P1053 residues. The bound peptide assumes a beta-hairpin conformation with a QRGPGR loop located at the very center of the binding pocket. The Fv and peptide surface areas buried upon binding are 601 A and 743 A(2), respectively, in the 0.5beta Fv-P1053 mean structure. The surface of P1053 interacting with the antibody is more extensive and the V3 peptide orientation in the binding site is significantly different compared with those derived from the crystal structures of a V3 peptide of the HIV-1 MN strain (V3(MN)) complexed to three different anti-peptide antibodies. The surface of P1053 that is in contact with the anti-protein antibody 0.5beta is likely to correspond to a solvent-exposed region in the native gp120 molecule. Some residues of this region of gp120 are involved in co-receptor binding, and in discrimination between different chemokine receptors utilized by the protein. Several highly variable residues in the V3 loop limit the specificity of the 0.5beta antibody, helping the virus to escape from the immune system. The highly conserved GPG sequence might have a role in maintaining the beta-hairpin conformation of the V3 loop despite insertions, deletions and mutations in the flanking regions.
Organizational Affiliation:
Department of Structural Biology, The Weizmann Institute of Science, Rehovot, 76100, Israel.