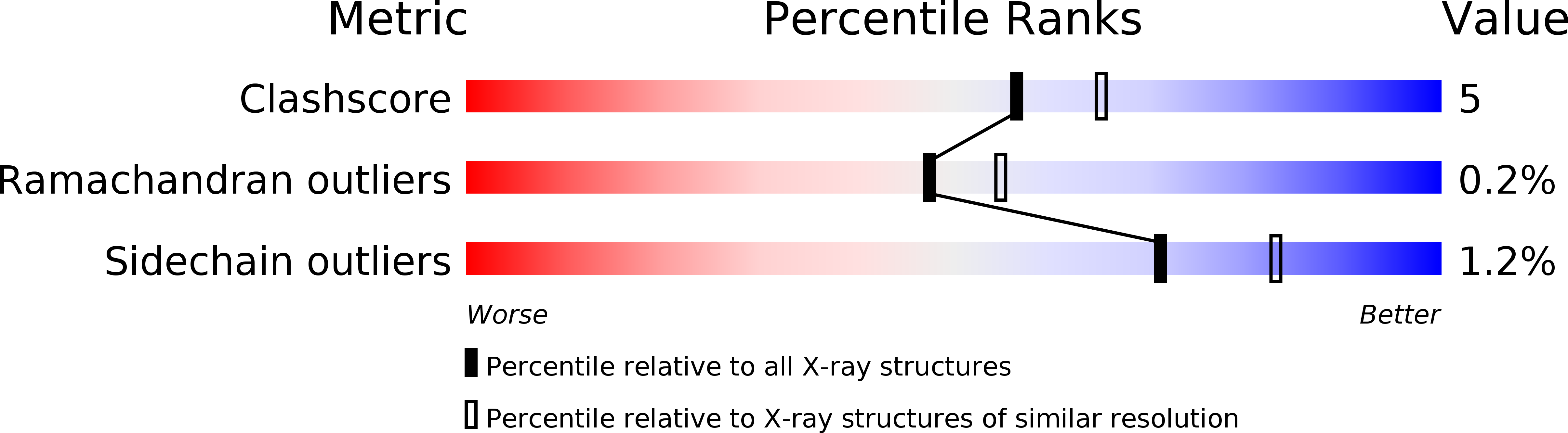
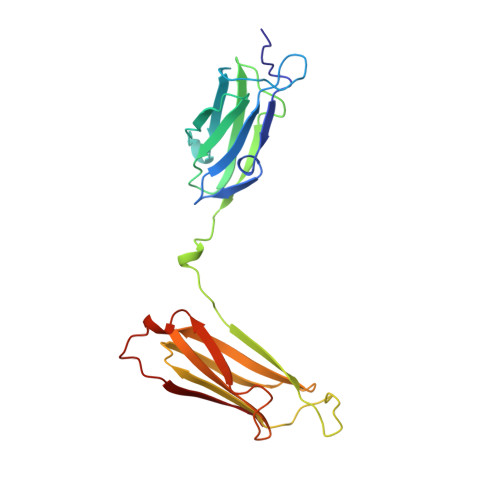
Structural basis for dimerization of the Dictyostelium gelation factor (ABP120) rod.
McCoy, A.J., Fucini, P., Noegel, A.A., Stewart, M.(1999) Nat Struct Biol 6: 836-841
- PubMed: 10467095
- DOI: https://doi.org/10.1038/12296
- Primary Citation of Related Structures:
1QFH - PubMed Abstract:
Gelation factor (ABP120) is one of the principal actin-cross-linking proteins of Dictyostelium discoideum. The extended molecule has an N-terminal 250-residue actin-binding domain and a rod constructed from six 100-residue repeats that have an Ig fold. The ability to dimerize is crucial to the actin cross-linking function of gelation factor and is mediated by the rod in which the two chains are arranged in an antiparallel fashion. We report the 2.2 A resolution crystal structure of rod domains 5 and 6, which shows that dimerization is mediated primarily by rod domain 6 and is the result of a double edge-to-edge extension of beta-sheets. Thus, contrary to earlier proposals, the chains of the dimeric gelation factor molecule overlap only within domain 6, and domains 1-5 do not pair with domains from the other chain. This information allows construction of a model of the gelation factor molecule and suggests how the chains in the related molecule filamin (ABP280) may interact.
Organizational Affiliation:
MRC Laboratory of Molecular Biology, Hills Rd, Cambridge CB2 2QH, UK.