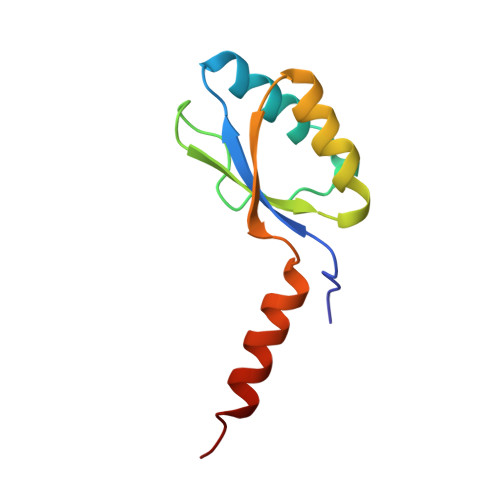
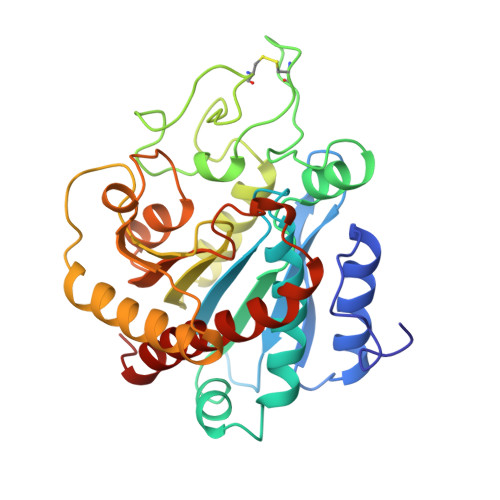
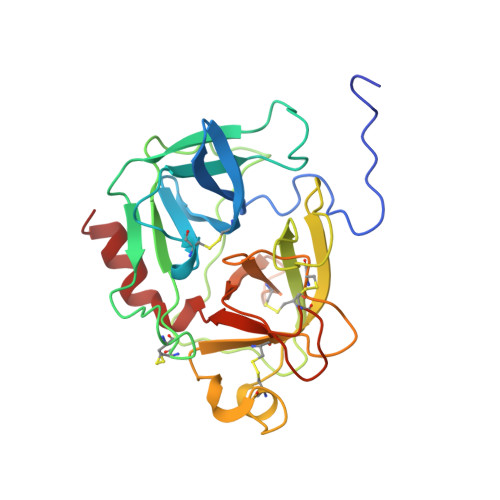
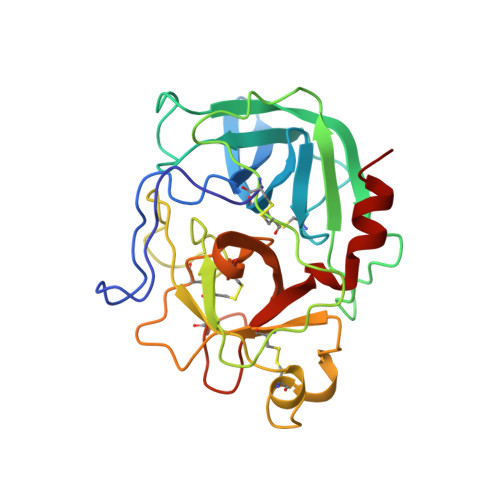
The three-dimensional structure of the native ternary complex of bovine pancreatic procarboxypeptidase A with proproteinase E and chymotrypsinogen C.
Gomis-Ruth, F.X., Gomez, M., Bode, W., Huber, R., Aviles, F.X.(1995) EMBO J 14: 4387-4394
- PubMed: 7556081
- DOI: https://doi.org/10.1002/j.1460-2075.1995.tb00117.x
- Primary Citation of Related Structures:
1PYT - PubMed Abstract:
The metalloexozymogen procarboxypeptidase A is mainly secreted in ruminants as a ternary complex with zymogens of two serine endoproteinases, chymotrypsinogen C and proproteinase E. The bovine complex has been crystallized, and its molecular structure analysed and refined at 2.6 A resolution to an R factor of 0.198. In this heterotrimer, the activation segment of procarboxypeptidase A essentially clamps the other two subunits, which shield the activation sites of the former from tryptic attack. In contrast, the propeptides of both serine proproteinases are freely accessible to trypsin. This arrangement explains the sequential and delayed activation of the constituent zymogens. Procarboxypeptidase A is virtually identical to the homologous monomeric porcine form. Chymotrypsinogen C displays structural features characteristic for chymotrypsins as well as elastases, except for its activation domain; similar to bovine chymotrypsinogen A, its binding site is not properly formed, while its surface located activation segment is disordered. The proproteinase E structure is fully ordered and strikingly similar to active porcine elastase; its specificity pocket is occluded, while the activation segment is fixed to the molecular surface. This first structure of a native zymogen from the proteinase E/elastase family does not fundamentally differ from the serine proproteinases known so far.
Organizational Affiliation:
Institut de Biologia Fonamental, Universitat Autònoma de Barcelona, Bellaterra, Spain.