The crystal structure of pneumococcal surface antigen PsaA reveals a metal-binding site and a novel structure for a putative ABC-type binding protein.
Lawrence, M.C., Pilling, P.A., Epa, V.C., Berry, A.M., Ogunniyi, A.D., Paton, J.C.(1998) Structure 6: 1553-1561
- PubMed: 9862808
- DOI: https://doi.org/10.1016/s0969-2126(98)00153-1
- Primary Citation of Related Structures:
1PSZ - PubMed Abstract:
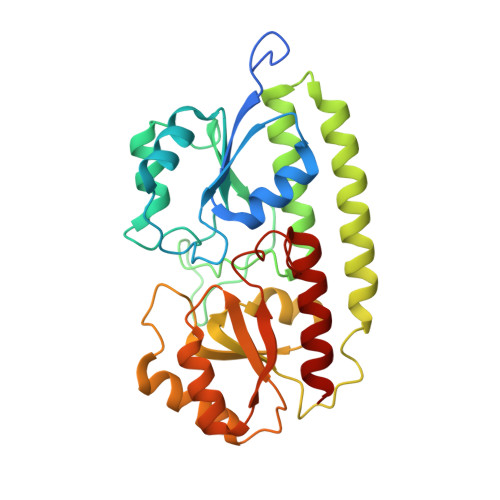
. The surface protein PsaA of the pathogenic bacterium Streptococcus pneumoniae plays an essential role in its virulence. PsaA is a putative ATP-binding cassette-type (ABC-type) binding protein involved in the uptake of Mn2+ and possibly Zn2+ and is considered to be both a potential drug target and and a candidate vaccine component. . The structure of PsaA has been determined to 2.0 A resolution using X-ray crystallography and is the first structure obtained for an ABC-type binding protein from a Gram-positive organism. The protein consists of two (beta/alpha)4 domains linked together by a single helix. A metal-binding site is formed in the domain interface by the sidechains of His67, His139, Glu205 and Asp280 and is occupied in the structure. . The structural topology of PsaA is fundamentally different from that of other ABC-type binding proteins determined thus far in that PsaA lacks the characteristic 'hinge peptides' involved in conformational change upon solute uptake and release. In our structure, the metal-binding site is probably occupied by Zn2+. The site seems to be well conserved amongst related receptors from both Gram-positive and Gram-negative bacteria.
Organizational Affiliation:
Biomolecular Research Institute 343 Royal Parade Parkville Victoria 3052 Australia. mike.lawrence@bioresi.com.au