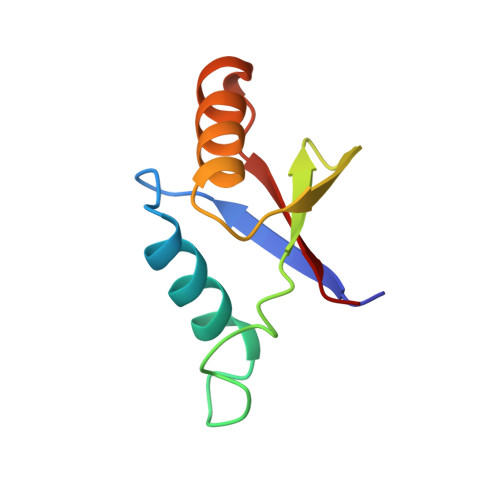
The solution structure of an N-terminally truncated version of the yeast CDC24p PB1 domain shows a different beta-sheet topology.
Leitner, D., Wahl, M., Labudde, D., Krause, G., Diehl, A., Schmieder, P., Pires, J.R., Fossi, M., Wiedemann, U., Leidert, M., Oschkinat, H.(2005) FEBS Lett 579: 3534-3538
- PubMed: 15961083
- DOI: https://doi.org/10.1016/j.febslet.2005.05.025
- Primary Citation of Related Structures:
1PQS - PubMed Abstract:
Phox and Bem1 (PB1) domains mediate protein-protein interactions via the formation of homo- or hetero-dimers. The C-terminal PB1 domain of yeast cell division cycle 24 (CDC24p), a guanine-nucleotide exchange factor involved in cell polarity establishment, is known to interact with the PB1 domain occurring in bud emergence MSB1 interacting 1 (BEM1p) during the regulation of the yeast budding process via its OPR/PC/AID (OPCA) motif. Here, we present the structure of an N-terminally truncated version of the Sc CDC24p PB1 domain. It shows a different topology of the beta-sheet than the long form. However, the C-terminal part of the structure shows the conserved PB1 domain features including the OPCA motif with a slight rearrangement of helix alpha1. Residues which are important for the heterodimerization with BEM1p are structurally preserved.
Organizational Affiliation:
Forschungsinstitut für Molekulare Pharmakologie, Robert-Rössle-Str. 10, 13125 Berlin, Germany. leitner@fmp-berlin.de