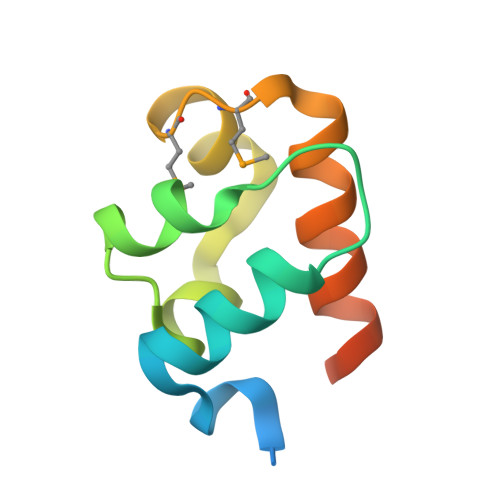
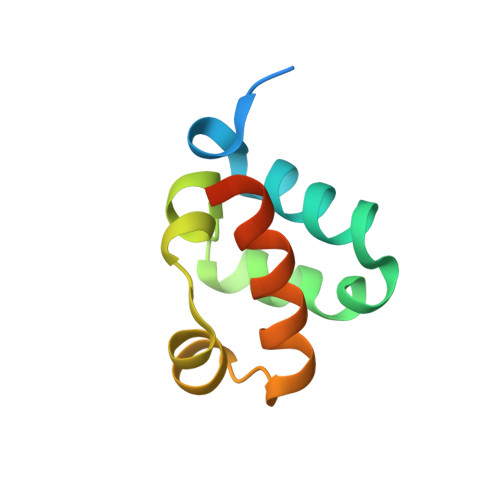
Structural organization of a Sex-comb-on-midleg/polyhomeotic copolymer.
Kim, C.A., Sawaya, M.R., Cascio, D., Kim, W., Bowie, J.U.(2005) J Biol Chem 280: 27769-27775
- PubMed: 15905166
- DOI: https://doi.org/10.1074/jbc.M503055200
- Primary Citation of Related Structures:
1PK1, 1PK3 - PubMed Abstract:
The polycomb group proteins are required for the stable maintenance of gene repression patterns established during development. They function as part of large multiprotein complexes created via a multitude of protein-protein interaction domains. Here we examine the interaction between the SAM domains of the polycomb group proteins polyhomeotic (Ph) and Sex-comb-on-midleg (Scm). Previously we showed that Ph-SAM polymerizes as a helical structure. We find that Scm-SAM also polymerizes, and a crystal structure reveals an architecture similar to the Ph-SAM polymer. These results suggest that Ph-SAM and Scm-SAM form a copolymer. Binding affinity measurements between Scm-SAM and Ph-SAM subunits in different orientations indicate a preference for the formation of a single junction copolymer. To provide a model of the copolymer, we determined the structure of the Ph-SAM/Scm-SAM junction. Similar binding modes are observed in both homo- and heterocomplex formation with minimal change in helix axis direction at the polymer joint. The copolymer model suggests that polymeric Scm complexes could extend beyond the local domains of polymeric Ph complexes on chromatin, possibly playing a role in long range repression.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of California, Los Angeles, California 90095, USA.