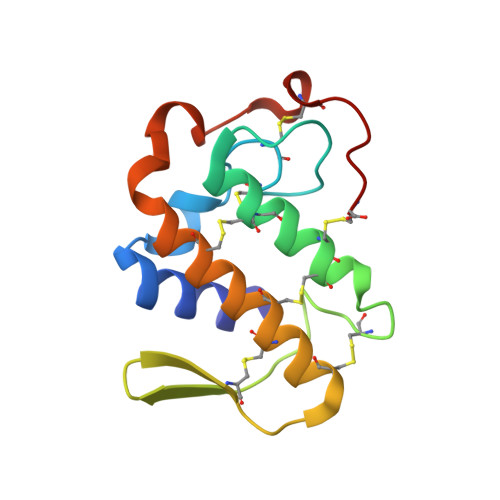
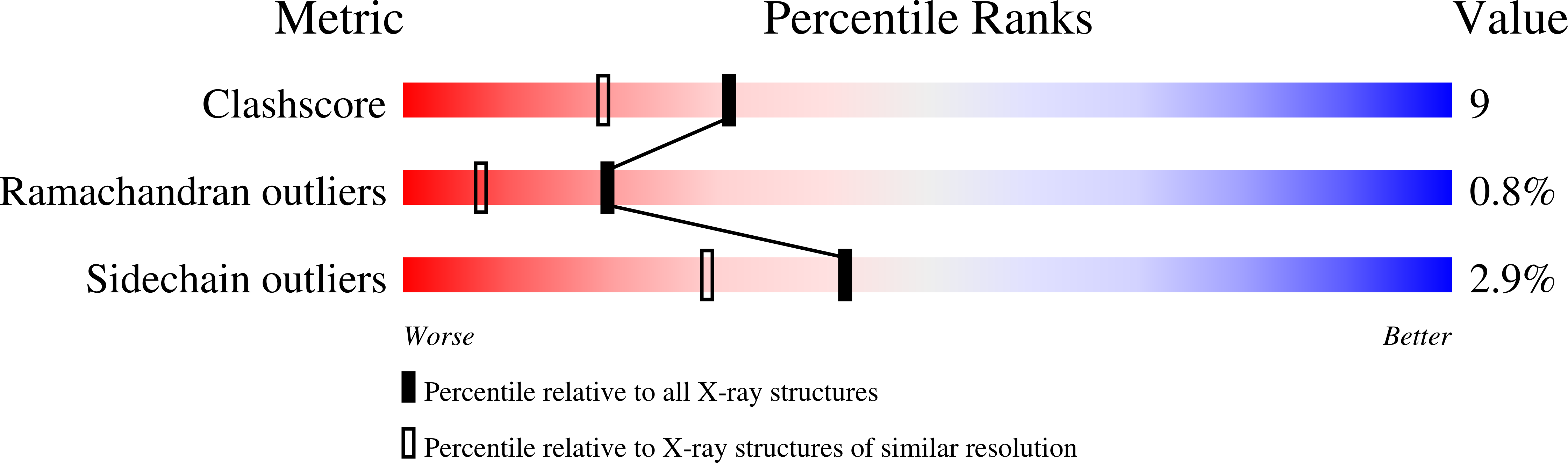Crystal structure of the complex of the secretory phospholipase A2 from Daboia russelli pulchella with an endogenic indole derivative, 2-carbamoylmethyl-5-propyl-octahydro-indol-7-yl-acetic acid at 1.8 A resolution.
Balasubramanya, R., Chandra, V., Kaur, P., Singh, T.P.(2005) Biochim Biophys Acta 1752: 177-185
- PubMed: 16122995
- DOI: https://doi.org/10.1016/j.bbapap.2005.07.020
- Primary Citation of Related Structures:
1OXL - PubMed Abstract:
Phospholipase A2 (PLA2) enzymes from snake venoms are approximately 14 kDa secretory proteins and catalyze the release of arachidonic acid which is the precursor of proinflammatory mediators such as prostaglandins, leukotrienes, thromboxanes and platelet-activating factors. The structure of the PLA2 enzyme purified from the venom of Daboia russelli pulchella was determined using molecular replacement method and refined to an R value of 18.3% for all the reflections to 1.8 A resolution. The structure contains two crystallographically independent molecules A and B which form an asymmetric homodimer. The Ca2+ ion was not detected in the present structure, however, a characteristic non-protein high quality electron density was observed at the substrate-binding site of molecule A which allowed a clear interpretation of a natural ligand identified as a derivative of indole, 2-carbamoylmethyl-5-propyl-octahydro-indol-7-yl)-acetic acid. The corresponding substrate-binding site in molecule B was empty. The ligand present in molecule A is involved in extensive interactions with the protein atoms including important catalytic residues such as Asp-49 and His-48. The results also show that the indole derivatives act as potent inhibitors of secretory group II PLA2 enzymes that can be further modified to be used as potential therapeutic agents.
Organizational Affiliation:
Department of Biophysics, All India Institute of Medical Sciences, New Delhi 110 029, India.