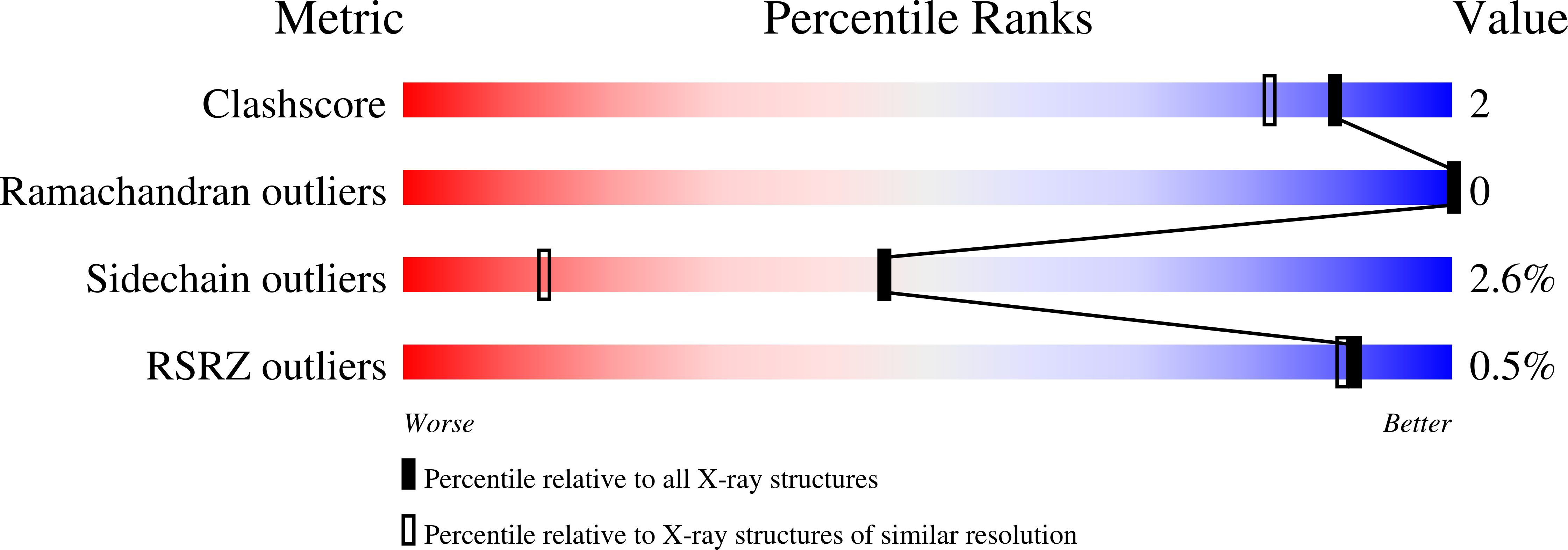
Crystal Structure of Truncated Human Beta-B1-Crystallin
Van Montfort, R.L.M., Bateman, O.A., Lubsen, N.H., Slingsby, C.(2003) Protein Sci 12: 2606
- PubMed: 14573871
- DOI: https://doi.org/10.1110/ps.03265903
- Primary Citation of Related Structures:
1OKI - PubMed Abstract:
Crystallins are long-lived proteins packed inside eye lens fiber cells that are essential in maintaining the transparency and refractive power of the eye lens. Members of the two-domain betagamma-crystallin family assemble into an array of oligomer sizes, forming intricate higher-order networks in the lens cell. Here we describe the 1.4 angstroms resolution crystal structure of a truncated version of human betaB1 that resembles an in vivo age-related truncation. The structure shows that unlike its close homolog, betaB2-crystallin, the homodimer is not domain swapped, but its domains are paired intramolecularly, as in more distantly related monomeric gamma-crystallins. However, the four-domain dimer resembles one half of the crystallographic bovine betaB2 tetramer and is similar to the engineered circular permuted rat betaB2. The crystal structure shows that the truncated betaB1 dimer is extremely well suited to form higher-order lattice interactions using its hydrophobic surface patches, linker regions, and sequence extensions.
Organizational Affiliation:
Department of Crystallography, Birkbeck College, London WC1E 7HX, UK.