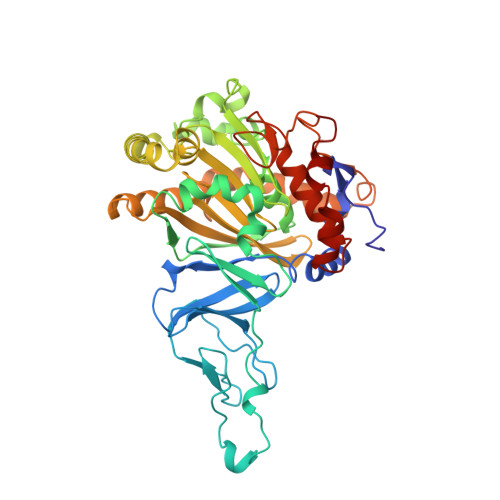
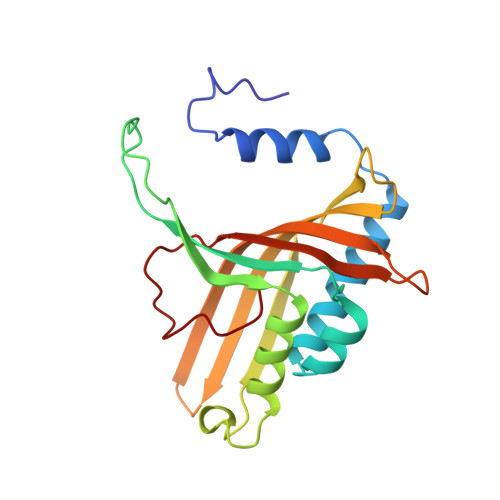
Structure of an aromatic-ring-hydroxylating dioxygenase-naphthalene 1,2-dioxygenase.
Kauppi, B., Lee, K., Carredano, E., Parales, R.E., Gibson, D.T., Eklund, H., Ramaswamy, S.(1998) Structure 6: 571-586
- PubMed: 9634695
- DOI: https://doi.org/10.1016/s0969-2126(98)00059-8
- Primary Citation of Related Structures:
1NDO - PubMed Abstract:
Pseudomonas sp. NCIB 9816-4 utilizes a multicomponent enzyme system to oxidize naphthalene to (+)-cis-(1R,2S)-dihydroxy-1,2-dihydronaphthalene. The enzyme component catalyzing this reaction, naphthalene 1,2-dioxygenase (NDO), belongs to a family of aromatic-ring-hydroxylating dioxygenases that oxidize aromatic hydrocarbons and related compounds to cis-arene diols. These enzymes utilize a mononuclear non-heme iron center to catalyze the addition of dioxygen to their respective substrates. The present study was conducted to provide essential structural information necessary for elucidating the mechanism of action of NDO. The three-dimensional structure of NDO has been determined at 2.25 A resolution. The molecule is an alpha 3 beta 3 hexamer. The alpha subunit has a beta-sheet domain that contains a Rieske [2Fe-2S] center and a catalytic domain that has a novel fold dominated by an antiparallel nine-stranded beta-pleated sheet against which helices pack. The active site contains a non-heme ferrous ion coordinated by His208, His213, Asp362 (bidentate) and a water molecule. Asn201 is positioned further away, 3.75 A, at the missing axial position of an octahedron. In the Rieske [2Fe-2S] center, one iron is coordinated by Cys81 and Cys101 and the other by His83 and His104. The domain structure and iron coordination of the Rieske domain is very similar to that of the cytochrome bc1 domain. The active-site iron center of one of the alpha subunits is directly connected by hydrogen bonds through a single amino acid, Asp205, to the Rieske [2Fe-2S] center in a neighboring alpha subunit. This is likely to be the main route for electron transfer.
Organizational Affiliation:
Department of Molecular Biology, Swedish University of Agricultural Sciences, Uppsala, Sweden.