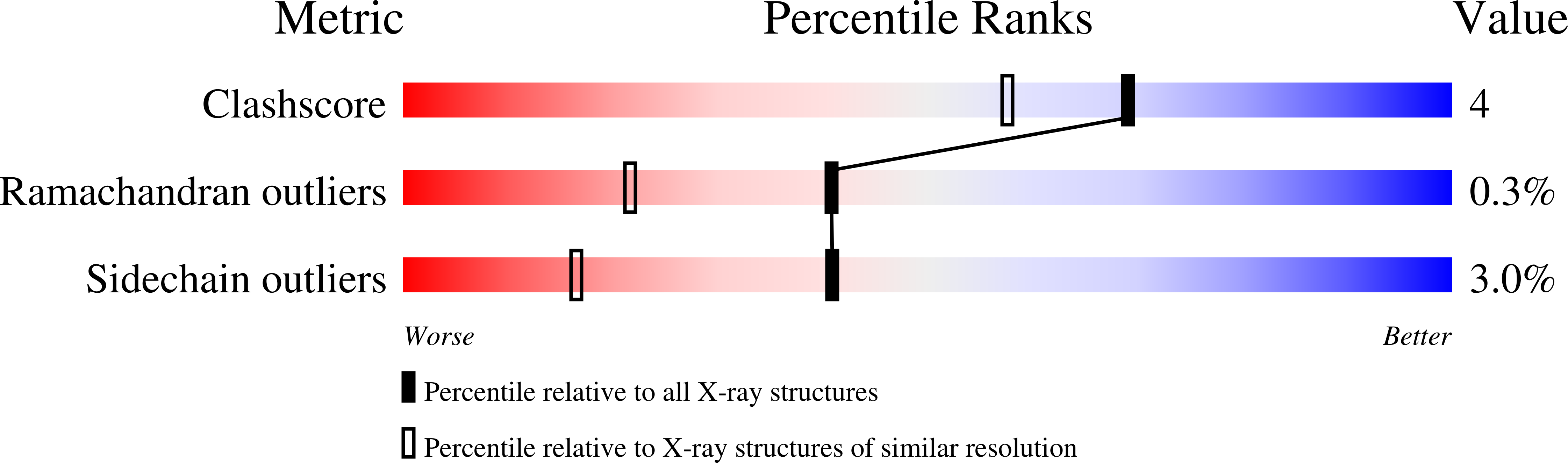The crystal structure of Escherichia coli MoaB suggests a probable role in molybdenum cofactor synthesis.
Sanishvili, R., Beasley, S., Skarina, T., Glesne, D., Joachimiak, A., Edwards, A., Savchenko, A.(2004) J Biol Chem 279: 42139-42146
- PubMed: 15269205
- DOI: https://doi.org/10.1074/jbc.M407694200
- Primary Citation of Related Structures:
1MKZ - PubMed Abstract:
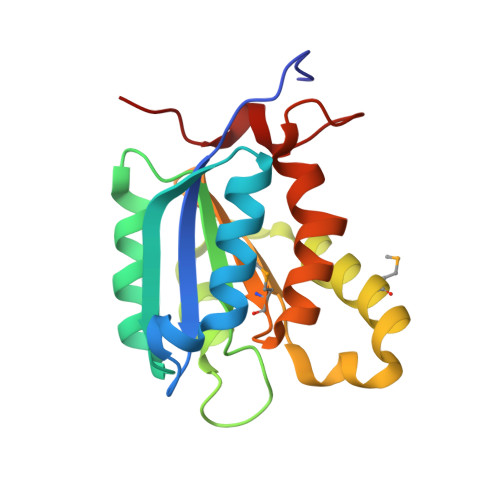
The crystal structure of Escherichia coli MoaB was determined by multiwavelength anomalous diffraction phasing and refined at 1.6-A resolution. The molecule displayed a modified Rossman fold. MoaB is assembled into a hexamer composed of two trimers. The monomers have high structural similarity with two proteins, MogA and MoeA, from the molybdenum cofactor synthesis pathway in E. coli, as well as with domains of mammalian gephyrin and plant Cnx1, which are also involved in molybdopterin synthesis. Structural comparison between these proteins and the amino acid conservation patterns revealed a putative active site in MoaB. The structural analysis of this site allowed to advance several hypothesis that can be tested in further studies.
Organizational Affiliation:
Biosciences, Structural Biology Center, Midwest Center for Sructural Genomics, Argonne National Laboratory, Argonne, Illinois 60439, USA.