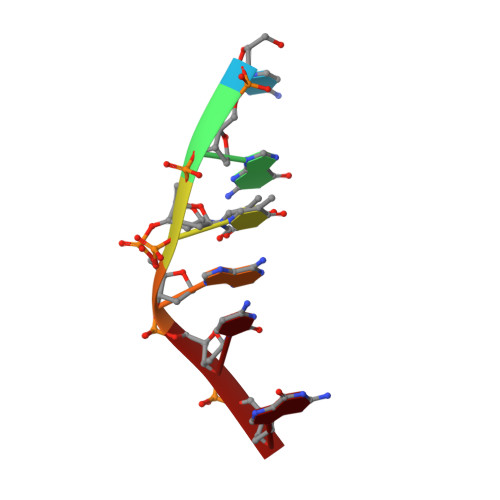
Heavy Going: the Atomic Resolution Structure of a Topoisomerase II Poison Intercalating into DNA in Multiple Orientations Also Reveals a Multiple Adenine-Thymine Hydrogen Bonding Pattern
Teixeira, S.C.M., Thorpe, J.H., Todd, A.K., Cardin, C.J.To be published.