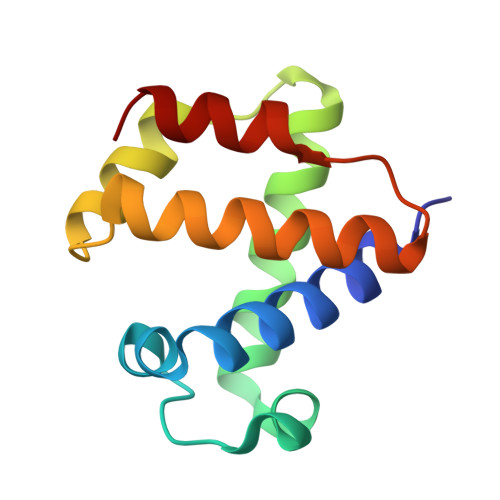
The 109 residue nerve tissue minihemoglobin from Cerebratulus lacteus highlights striking structural plasticity of the alpha-helical globin fold
Pesce, A., Nardini, M., Dewilde, S., Geuens, E., Yamauchi, k., Ascenzi, P., Riggs, A.F., Moens, L., Bolognesi, M.(2002) Structure 10: 725-735
- PubMed: 12015154
- DOI: https://doi.org/10.1016/s0969-2126(02)00763-3
- Primary Citation of Related Structures:
1KR7 - PubMed Abstract:
A very short hemoglobin (CerHb; 109 amino acids) binds O(2) cooperatively in the nerve tissue of the nemertean worm Cerebratulus lacteus to sustain neural activity during anoxia. Sequence analysis suggests that CerHb tertiary structure may be unique among the known globin fold evolutionary variants. The X-ray structure of oxygenated CerHb (R factor 15.3%, at 1.5 A resolution) displays deletion of the globin N-terminal A helix, an extended GH region, a very short H helix, and heme solvent shielding based on specific aromatic residues. The heme-bound O(2) is stabilized by hydrogen bonds to the distal TyrB10-GlnE7 pair. Ligand access to heme may take place through a wide protein matrix tunnel connecting the distal site to a surface cleft located between the E and H helices.
Organizational Affiliation:
Department of Physics, INFM, Advanced Biotechnology Centre, University of Genova, Largo Rosanna Benzi 10, I-16146 Genova, Italy.