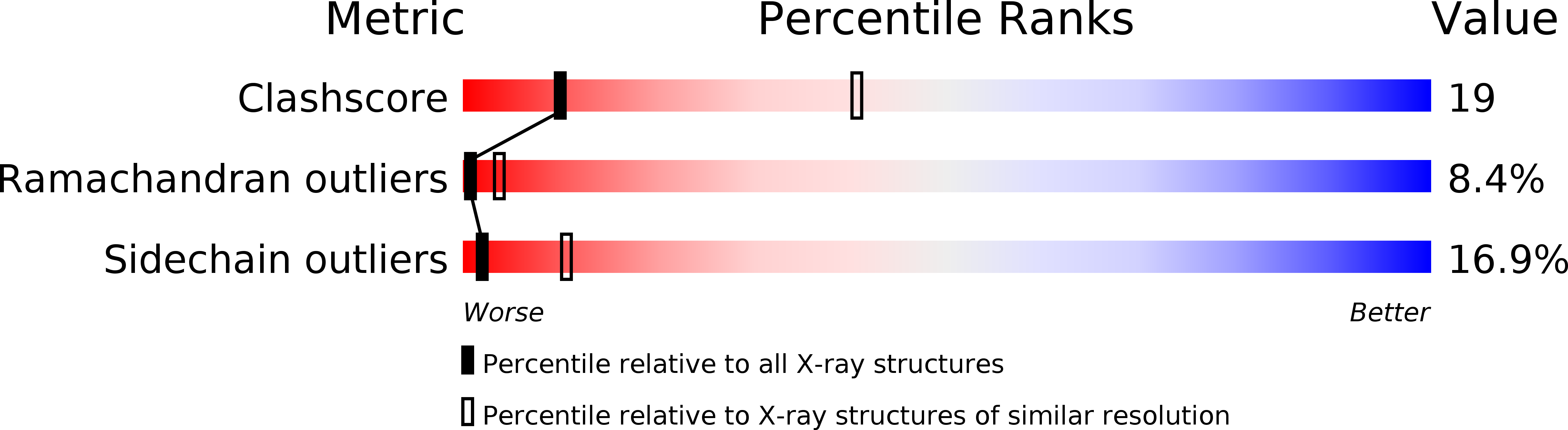Unexpected binding mode of tick anticoagulant peptide complexed to bovine factor Xa.
Wei, A., Alexander, R.S., Duke, J., Ross, H., Rosenfeld, S.A., Chang, C.H.(1998) J Mol Biol 283: 147-154
- PubMed: 9761680
- DOI: https://doi.org/10.1006/jmbi.1998.2069
- Primary Citation of Related Structures:
1KIG - PubMed Abstract:
The structure of recombinant tick anticoagulant peptide (rTAP) complexed to bovine factor Xa at 3.0 A resolution reveals the structural basis for the specificity and the high affinity of rTAP. Three N-terminal residues, Tyr501, Asn502 and Arg503, play a critical role in the complex formation as suggested by earlier mutagenic studies and the ornithodorin-thrombin complex. Unexpectedly, the side-chain of Tyr501 is located in the S1 pocket, although factor Xa favors arginine as a P1 residue. Arg503 is located at the aryl binding pocket and forms a salt-bridge with Glu97 of factor Xa. The autolysis loop, which is disordered in the uninhibited factor Xa structure, is involved in the formation of the complex as a part of the secondary binding site. The C-terminal helix of rTAP interacts with factor Xa as a secondary binding determinant. The N-terminal residues of rTAP reorganize during the formation of the factor Xa-rTAP complex from the conformation found in the solution into an extended conformation. The presence of the secondary binding site confirms the proposed two-step kinetic mechanism based on the results of a mutagenesis study.
Organizational Affiliation:
Department of Chemical and Physical Sciences, The DuPont Pharmaceuticals Company, Wilmington DE 19880, USA.