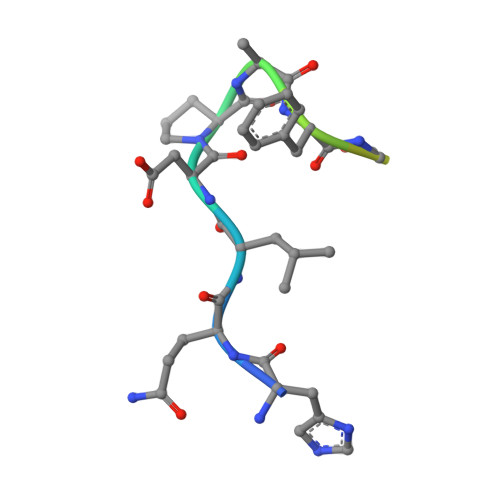
Epitope recognition by diverse antibodies suggests conformational convergence in an antibody response.
Nair, D.T., Singh, K., Siddiqui, Z., Nayak, B.P., Rao, K.V., Salunke, D.M.(2002) J Immunol 168: 2371-2382
- PubMed: 11859128
- DOI: https://doi.org/10.4049/jimmunol.168.5.2371
- Primary Citation of Related Structures:
1KC5, 1KCS, 1KCU, 1KCV - PubMed Abstract:
Crystal structures of distinct mAbs that recognize a common epitope of a peptide Ag have been determined and analyzed in the unbound and bound forms. These Abs display dissimilar binding site structures in the absence of the Ag. The dissimilarity is primarily expressed in the conformations of complementarity-determining region H3, which is responsible for defining the epitope specificity. Interestingly, however, the three Abs exhibit similar complementarity-determining region conformations in the Ag binding site while recognizing the common epitope, indicating that different pathways of binding are used for Ag recognition. The epitope also exhibits conformational similarity when bound to each of these Abs, although the peptide Ag was otherwise flexible. The observed conformational convergence in the epitope and the Ag binding site was facilitated by the plasticity in the nature of interactions.
Organizational Affiliation:
Structural Biology Unit, National Institute of Immunology, New Delhi 110067, India.