Molecular structure of kanamycin nucleotidyltransferase determined to 3.0-A resolution.
Sakon, J., Liao, H.H., Kanikula, A.M., Benning, M.M., Rayment, I., Holden, H.M.(1993) Biochemistry 32: 11977-11984
- PubMed: 8218273
- DOI: https://doi.org/10.1021/bi00096a006
- Primary Citation of Related Structures:
1KAN - PubMed Abstract:
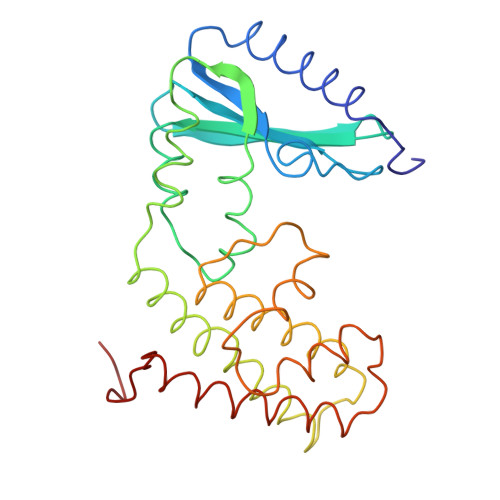
Kanamycin nucleotidyltransferase, as originally isolated from Staphylococcus aureus, inactivates the antibiotic kanamycin by catalyzing the transfer of a nucleotidyl group from nucleoside triphosphates such as ATP to the 4'-hydroxyl group of the aminoglycoside. The molecular structure of the enzyme described here was determined by X-ray crystallographic analysis to a resolution of 3.0 A. Crystals employed in the investigation belonged to the space group P4(3)2(1)2 with unit cell dimensions of a = b = 78.9 A and c = 219.2 A. An electron density map phased with seven heavy-atom derivatives revealed that the molecules packed in the crystalline lattice as dimers exhibiting local 2-fold rotation axes. Subsequent symmetry averaging and solvent flattening improved the quality of the electron density such that it was possible to completely trace the 253 amino acid polypeptide chain. Each monomer is divided into two distinct structural domains: the N-terminal motif composed of residues Met 1-Glu 127 and the C-terminal half delineated by residues Ala 128-Phe 253. The N-terminal region is characterized by a five-stranded mixed beta-pleated sheet whereas the C-terminal domain contains five alpha-helices, four of which form an up-and-down alpha-helical bundle very similar to that observed in cytochrome c'. The two subunits wrap about one another to form an ellipsoid with a pronounced cleft that could easily accommodate the various aminoglycosides known to bind to the enzyme.
Organizational Affiliation:
Institute for Enzyme Research, Graduate School, University of Wisconsin, Madison 53705.