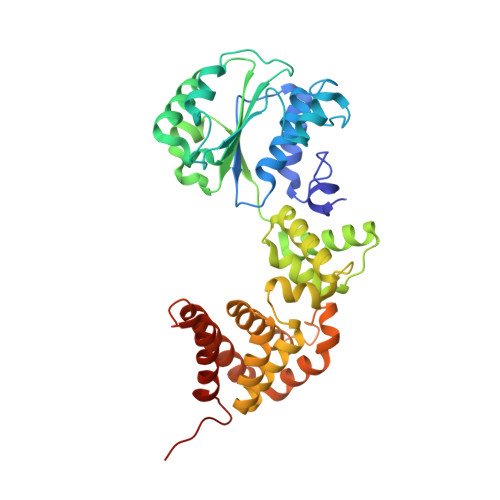
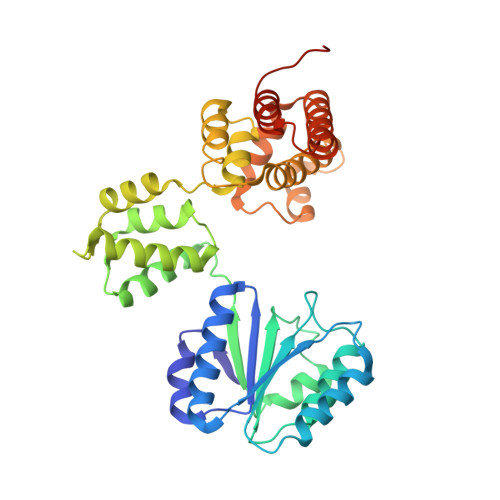
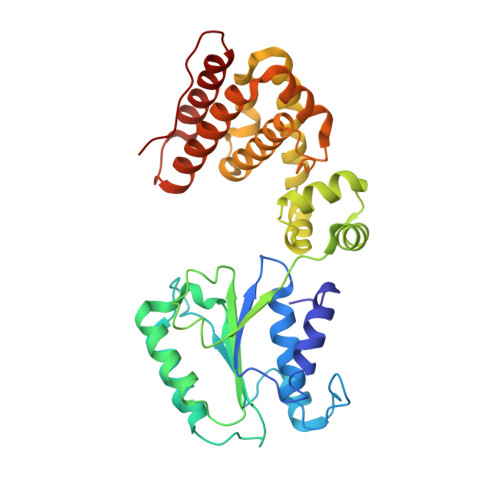
Crystal structure of the processivity clamp loader gamma (gamma) complex of E. coli DNA polymerase III.
Jeruzalmi, D., O'Donnell, M., Kuriyan, J.(2001) Cell 106: 429-441
- PubMed: 11525729
- DOI: https://doi.org/10.1016/s0092-8674(01)00463-9
- Primary Citation of Related Structures:
1JR3 - PubMed Abstract:
The gamma complex, an AAA+ ATPase, is the bacterial homolog of eukaryotic replication factor C (RFC) that loads the sliding clamp (beta, homologous to PCNA) onto DNA. The 2.7/3.0 A crystal structure of gamma complex reveals a pentameric arrangement of subunits, with stoichiometry delta':gamma(3):delta. The C-terminal domains of the subunits form a circular collar that supports an asymmetric arrangement of the N-terminal ATP binding domains of the gamma motor and the structurally related domains of the delta' stator and the delta wrench. The structure suggests a mechanism by which the gamma complex switches between a closed state, in which the beta-interacting element of delta is hidden by delta', and an open form similar to the crystal structure, in which delta is free to bind to beta.
Organizational Affiliation:
Howard Hughes Medical Institute, The Rockefeller University, 1230 York Avenue, New York, NY 10021, USA.