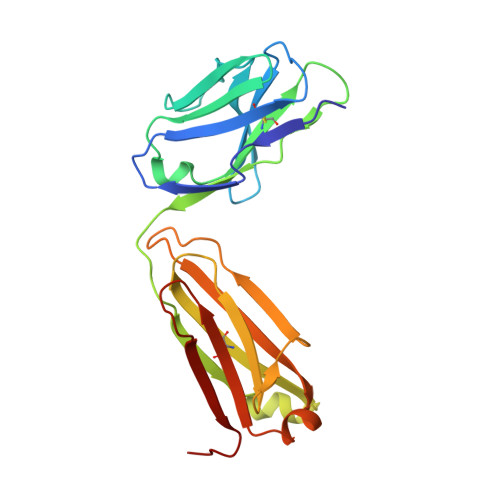
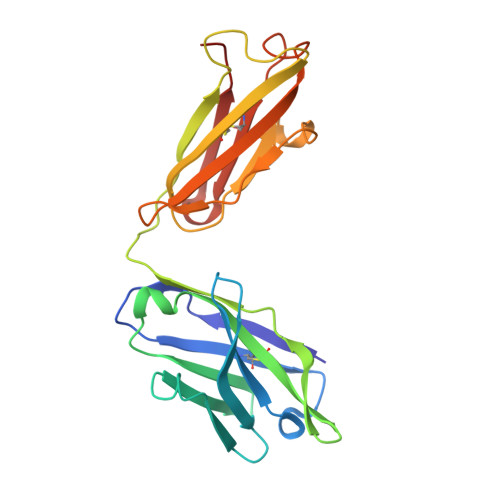
Structural analysis of mutants of high-affinity and low-affinity p-azophenylarsonate-specific antibodies generated by alanine scanning of heavy chain complementarity-determining region 2.
Parhami-Seren, B., Viswanathan, M., Strong, R.K., Margolies, M.N.(2001) J Immunol 167: 5129-5135
- PubMed: 11673524
- DOI: https://doi.org/10.4049/jimmunol.167.9.5129
- Primary Citation of Related Structures:
1JFQ - PubMed Abstract:
Alanine scanning was used to determine the affinity contributions of 10 side chain amino acids (residues at position 50-60 inclusive) of H chain complementarity-determining region 2 (HCDR2) of the somatically mutated high-affinity anti-p-azophenylarsonate Ab, 36-71. Each mutated H chain gene was expressed in the context of mutated (36-71L) and the unmutated (36-65L) L chains to also assess the contribution of L chain mutations to affinity. Combined data from fluorescence quenching, direct binding, inhibition, and capture assays indicated that mutating H:Tyr(50) and H:Tyr(57) to Ala in the 36-71 H chain results in significant loss of binding with both mutated (36-71L) or unmutated (36-65L) L chain, although the decrease was more pronounced when unmutated L chain was used. All other HCDR2 mutations in 36-71 had minimal effect on Ab affinity when expressed with 36-71 L chain. However, in the context of unmutated L chain, of H:Gly(54) to Ala resulted in significant loss of binding, while Abs containing Asn(52) to Ala, Pro(53) to Ala, or Ile(58) to Ala mutation exhibited 4.3- to 7.1-fold reduced affinities. When alanine scanning was performed instead on certain HCDR2 residues of the germline-encoded (unmutated) 36-65 Ab and expressed with unmutated L chain as Fab in bacteria, these mutants exhibited affinities similar to or slightly higher than the wild-type 36-65. These findings indicate an important role of certain HCDR2 side chain residues on Ab affinity and the constraints imposed by L chain mutations in maintaining Ag binding.
Organizational Affiliation:
Department of Biochemistry, College of Medicine, University of Vermont, Burlington, VT 05405, USA. bparhami@zoo.uvm.edu