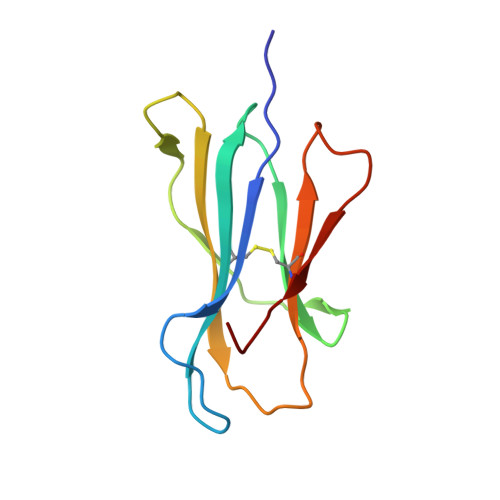
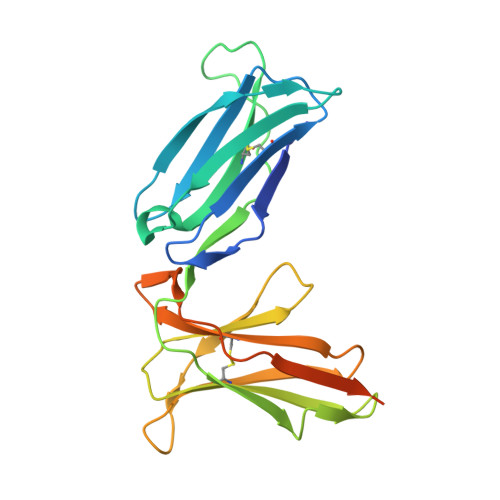
Crystal structure of the human natural killer cell inhibitory receptor KIR2DL1-HLA-Cw4 complex.
Fan, Q.R., Long, E.O., Wiley, D.C.(2001) Nat Immunol 2: 452-460
- PubMed: 11323700
- DOI: https://doi.org/10.1038/87766
- Primary Citation of Related Structures:
1IM9 - PubMed Abstract:
Inhibitory natural killer (NK) cell receptors down-regulate the cytotoxicity of NK cells upon recognition of specific class I major histocompatibility complex (MHC) molecules on target cells. We report here the crystal structure of the inhibitory human killer cell immunoglobulin-like receptor 2DL1 (KIR2DL1) bound to its class I MHC ligand, HLA-Cw4. The KIR2DL1-HLA-Cw4 interface exhibits charge and shape complementarity. Specificity is mediated by a pocket in KIR2DL1 that hosts the Lys80 residue of HLA-Cw4. Many residues conserved in HLA-C and in KIR2DL receptors make different interactions in KIR2DL1-HLA-Cw4 and in a previously reported KIR2DL2-HLA-Cw3 complex. A dimeric aggregate of KIR-HLA-C complexes was observed in one KIR2DL1-HLA-Cw4 crystal. Most of the amino acids that differ between human and chimpanzee KIRs with HLA-C specificities form solvent-accessible clusters outside the KIR-HLA interface, which suggests undiscovered interactions by KIRs.
Organizational Affiliation:
Department of Molecular and Cellular Biology and Howard Hughes Medical Institute, Harvard University, 7 Divinity Avenue, Cambridge, MA 02138, USA.