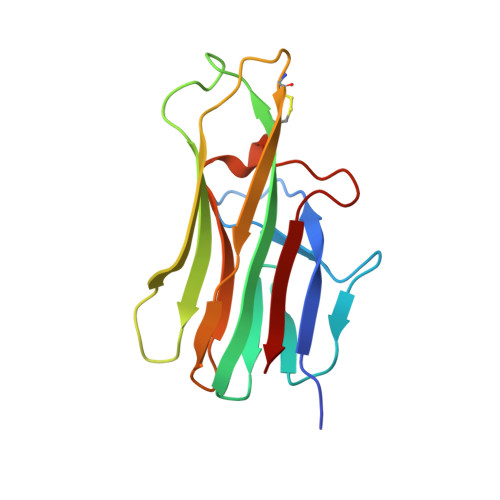
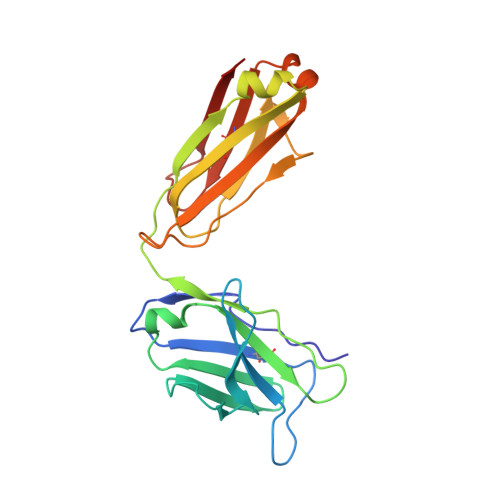
Structure of CD40 ligand in complex with the Fab fragment of a neutralizing humanized antibody.
Karpusas, M., Lucci, J., Ferrant, J., Benjamin, C., Taylor, F.R., Strauch, K., Garber, E., Hsu, Y.M.(2001) Structure 9: 321-329
- PubMed: 11525169
- DOI: https://doi.org/10.1016/s0969-2126(01)00590-1
- Primary Citation of Related Structures:
1I9R - PubMed Abstract:
CD40 ligand (CD40L or CD154), a member of the tumor necrosis factor (TNF) family, plays a critical role in both humoral and cellular immune responses and has been implicated in biological pathways involving epithelial cells, fibroblasts, and platelets. Such a pathway is T cell-mediated B cell activation, a process that occurs through the interaction of CD40L with CD40 receptor expressed on B cells. It results in various B cell responses, including immunoglobulin isotype switching and B cell differentiation and proliferation. These responses can be inhibited by the monoclonal antibody 5c8, which binds with high affinity to CD40L.
Organizational Affiliation:
Biogen, Inc, Cambridge, Massachusetts 02142, USA. michael_karpusas@biogen.com.